Author: Jason Brownlee
Random forest is an ensemble machine learning algorithm.
It is perhaps the most popular and widely used machine learning algorithm given its good or excellent performance across a wide range of classification and regression predictive modeling problems.
It is also easy to use given that it has few key hyperparameters and sensible heuristics for configuring these hyperparameters.
In this tutorial, you will discover how to develop a random forest ensemble for classification and regression.
After completing this tutorial, you will know:
- Random forest ensemble is an ensemble of decision trees and a natural extension of bagging.
- How to use the random forest ensemble for classification and regression with scikit-learn.
- How to explore the effect of random forest model hyperparameters on model performance.
Let’s get started.

How to Develop a Random Forest Ensemble in Python
Photo by Sheila Sund, some rights reserved.
Tutorial Overview
This tutorial is divided into three parts; they are:
- Random Forest Algorithm
- Random Forest Scikit-Learn API
- Random Forest for Classification
- Random Forest for Regression
- Random Forest Hyperparameters
- Explore Number of Samples
- Explore Number of Features
- Explore Number of Trees
- Explore Tree Depth
Random Forest Algorithm
Random forest is an ensemble of decision tree algorithms.
It is an extension of bootstrap aggregation (bagging) of decision trees and can be used for classification and regression problems.
In bagging, a number of decision trees are created where each tree is created from a different bootstrap sample of the training dataset. A bootstrap sample is a sample of the training dataset where a sample may appear more than once in the sample, referred to as sampling with replacement.
Bagging is an effective ensemble algorithm as each decision tree is fit on a slightly different training dataset, and in turn, has a slightly different performance. Unlike normal decision tree models, such as classification and regression trees (CART), trees used in the ensemble are unpruned, making them slightly overfit to the training dataset. This is desirable as it helps to make each tree more different and have less correlated predictions or prediction errors.
Predictions from the trees are averaged across all decision trees resulting in better performance than any single tree in the model.
Each model in the ensemble is then used to generate a prediction for a new sample and these m predictions are averaged to give the forest’s prediction
— Page 199, Applied Predictive Modeling, 2013.
A prediction on a regression problem is the average of the prediction across the trees in the ensemble. A prediction on a classification problem is the majority vote for the class label across the trees in the ensemble.
- Regression: Prediction is the average prediction across the decision trees.
- Classification: Prediction is the majority vote class label predicted across the decision trees.
As with bagging, each tree in the forest casts a vote for the classification of a new sample, and the proportion of votes in each class across the ensemble is the predicted probability vector.
— Page 387, Applied Predictive Modeling, 2013.
Random forest involves constructing a large number of decision trees from bootstrap samples from the training dataset, like bagging.
Unlike bagging, random forest also involves selecting a subset of input features (columns or variables) at each split point in the construction of trees. Typically, constructing a decision tree involves evaluating the value for each input variable in the data in order to select a split point. By reducing the features to a random subset that may be considered at each split point, it forces each decision tree in the ensemble to be more different.
Random forests provide an improvement over bagged trees by way of a small tweak that decorrelates the trees. […] But when building these decision trees, each time a split in a tree is considered, a random sample of m predictors is chosen as split candidates from the full set of p predictors.
— Page 320, An Introduction to Statistical Learning with Applications in R, 2014.
The effect is that the predictions, and in turn, prediction errors, made by each tree in the ensemble are more different or less correlated. When the predictions from these less correlated trees are averaged to make a prediction, it often results in better performance than bagged decision trees.
Perhaps the most important hyperparameter to tune for the random forest is the number of random features to consider at each split point.
Random forests’ tuning parameter is the number of randomly selected predictors, k, to choose from at each split, and is commonly referred to as mtry. In the regression context, Breiman (2001) recommends setting mtry to be one-third of the number of predictors.
— Page 199, Applied Predictive Modeling, 2013.
A good heuristic for regression is to set this hyperparameter to 1/3 the number of input features.
- num_features_for_split = total_input_features / 3
For classification problems, Breiman (2001) recommends setting mtry to the square root of the number of predictors.
— Page 387, Applied Predictive Modeling, 2013.
A good heuristic for classification is to set this hyperparameter to the square root of the number of input features.
- num_features_for_split = sqrt(total_input_features)
Another important hyperparameter to tune is the depth of the decision trees. Deeper trees are often more overfit to the training data, but also less correlated, which in turn may improve the performance of the ensemble. Depths from 1 to 10 levels may be effective.
Finally, the number of decision trees in the ensemble can be set. Often, this is increased until no further improvement is seen.
Random Forest Scikit-Learn API
Random Forest ensembles can be implemented from scratch, although this can be challenging for beginners.
The scikit-learn Python machine learning library provides an implementation of Random Forest for machine learning.
It is available in modern versions of the library.
First, confirm that you are using a modern version of the library by running the following script:
# check scikit-learn version import sklearn print(sklearn.__version__)
Running the script will print your version of scikit-learn.
Your version should be the same or higher. If not, you must upgrade your version of the scikit-learn library.
0.22.1
Random Forest is provided via the RandomForestRegressor and RandomForestClassifier classes.
Both models operate the same way and take the same arguments that influence how the decision trees are created.
Randomness is used in the construction of the model. This means that each time the algorithm is run on the same data, it will produce a slightly different model.
When using machine learning algorithms that have a stochastic learning algorithm, it is good practice to evaluate them by averaging their performance across multiple runs or repeats of cross-validation. When fitting a final model, it may be desirable to either increase the number of trees until the variance of the model is reduced across repeated evaluations, or to fit multiple final models and average their predictions.
Let’s take a look at how to develop a Random Forest ensemble for both classification and regression tasks.
Random Forest for Classification
In this section, we will look at using Random Forest for a classification problem.
First, we can use the make_classification() function to create a synthetic binary classification problem with 1,000 examples and 20 input features.
The complete example is listed below.
# test classification dataset from sklearn.datasets import make_classification # define dataset X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=3) # summarize the dataset print(X.shape, y.shape)
Running the example creates the dataset and summarizes the shape of the input and output components.
(1000, 20) (1000,)
Next, we can evaluate a random forest algorithm on this dataset.
We will evaluate the model using repeated stratified k-fold cross-validation, with three repeats and 10 folds. We will report the mean and standard deviation of the accuracy of the model across all repeats and folds.
# evaluate random forest algorithm for classification
from numpy import mean
from numpy import std
from sklearn.datasets import make_classification
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.ensemble import RandomForestClassifier
# define dataset
X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=3)
# define the model
model = RandomForestClassifier()
# evaluate the model
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
# report performance
print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores)))
Running the example reports the mean and standard deviation accuracy of the model.
Your specific results may vary given the stochastic nature of the learning algorithm. Try running the example a few times.
In this case, we can see the random forest ensemble with default hyperparameters achieves a classification accuracy of about 90.5 percent.
Accuracy: 0.905 (0.025)
We can also use the random forest model as a final model and make predictions for classification.
First, the random forest ensemble is fit on all available data, then the predict() function can be called to make predictions on new data.
The example below demonstrates this on our binary classification dataset.
# make predictions using random forest for classification
from sklearn.datasets import make_classification
from sklearn.ensemble import RandomForestClassifier
# define dataset
X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=3)
# define the model
model = RandomForestClassifier()
# fit the model on the whole dataset
model.fit(X, y)
# make a single prediction
row = [[-8.52381793,5.24451077,-12.14967704,-2.92949242,0.99314133,0.67326595,-0.38657932,1.27955683,-0.60712621,3.20807316,0.60504151,-1.38706415,8.92444588,-7.43027595,-2.33653219,1.10358169,0.21547782,1.05057966,0.6975331,0.26076035]]
yhat = model.predict(row)
print('Predicted Class: %d' % yhat[0])
Running the example fits the random forest ensemble model on the entire dataset and is then used to make a prediction on a new row of data, as we might when using the model in an application.
Predicted Class: 0
Now that we are familiar with using random forest for classification, let’s look at the API for regression.
Random Forest for Regression
In this section, we will look at using random forests for a regression problem.
First, we can use the make_regression() function to create a synthetic regression problem with 1,000 examples and 20 input features.
The complete example is listed below.
# test regression dataset from sklearn.datasets import make_regression # define dataset X, y = make_regression(n_samples=1000, n_features=20, n_informative=15, noise=0.1, random_state=2) # summarize the dataset print(X.shape, y.shape)
Running the example creates the dataset and summarizes the shape of the input and output components.
(1000, 20) (1000,)
Next, we can evaluate a random forest algorithm on this dataset.
As we did with the last section, we will evaluate the model using repeated k-fold cross-validation, with three repeats and 10 folds. We will report the mean absolute error (MAE) of the model across all repeats and folds. The scikit-learn library makes the MAE negative so that it is maximized instead of minimized. This means that larger negative MAE are better and a perfect model has a MAE of 0.
The complete example is listed below.
# evaluate random forest ensemble for regression
from numpy import mean
from numpy import std
from sklearn.datasets import make_regression
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedKFold
from sklearn.ensemble import RandomForestRegressor
# define dataset
X, y = make_regression(n_samples=1000, n_features=20, n_informative=15, noise=0.1, random_state=2)
# define the model
model = RandomForestRegressor()
# evaluate the model
cv = RepeatedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(model, X, y, scoring='neg_mean_absolute_error', cv=cv, n_jobs=-1, error_score='raise')
# report performance
print('MAE: %.3f (%.3f)' % (mean(n_scores), std(n_scores)))
Running the example reports the mean and standard deviation accuracy of the model.
Your specific results may vary given the stochastic nature of the learning algorithm. Try running the example a few times.
In this case, we can see the random forest ensemble with default hyperparameters achieves a MAE of about 90.
MAE: -90.149 (7.924)
We can also use the random forest model as a final model and make predictions for regression.
First, the random forest ensemble is fit on all available data, then the predict() function can be called to make predictions on new data.
The example below demonstrates this on our regression dataset.
# random forest for making predictions for regression
from sklearn.datasets import make_regression
from sklearn.ensemble import RandomForestRegressor
# define dataset
X, y = make_regression(n_samples=1000, n_features=20, n_informative=15, noise=0.1, random_state=2)
# define the model
model = RandomForestRegressor()
# fit the model on the whole dataset
model.fit(X, y)
# make a single prediction
row = [[-0.89483109,-1.0670149,-0.25448694,-0.53850126,0.21082105,1.37435592,0.71203659,0.73093031,-1.25878104,-2.01656886,0.51906798,0.62767387,0.96250155,1.31410617,-1.25527295,-0.85079036,0.24129757,-0.17571721,-1.11454339,0.36268268]]
yhat = model.predict(row)
print('Prediction: %d' % yhat[0])
Running the example fits the random forest ensemble model on the entire dataset and is then used to make a prediction on a new row of data, as we might when using the model in an application.
Prediction: -173
Now that we are familiar with using the scikit-learn API to evaluate and use random forest ensembles, let’s look at configuring the model.
Random Forest Hyperparameters
In this section, we will take a closer look at some of the hyperparameters you should consider tuning for the random forest ensemble and their effect on model performance.
Explore Number of Samples
Each decision tree in the ensemble is fit on a bootstrap sample drawn from the training dataset.
This can be turned off by setting the “bootstrap” argument to False, if you desire. In that case, the whole training dataset will be used to train each decision tree. This is not recommended.
The “max_samples” argument can be set to a float between 0 and 1 to control the percentage of the size of the training dataset to make the bootstrap sample used to train each decision tree.
For example, if the training dataset has 100 rows, the max_samples argument could be set to 0.5 and each decision tree will be fit on a bootstrap sample with (100 * 0.5) or 50 rows of data.
A smaller sample size will make trees more different, and a larger sample size will make the trees more similar. Setting max_samples to “None” will make the sample size the same size as the training dataset and this is the default.
The example below demonstrates the effect of different bootstrap sample sizes from 10 percent to 100 percent on the random forest algorithm.
# explore random forest bootstrap sample size on performance
from numpy import mean
from numpy import std
from sklearn.datasets import make_classification
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.ensemble import RandomForestClassifier
from matplotlib import pyplot
# get the dataset
def get_dataset():
X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=3)
return X, y
# get a list of models to evaluate
def get_models():
models = dict()
models['10'] = RandomForestClassifier(max_samples=0.1)
models['20'] = RandomForestClassifier(max_samples=0.2)
models['30'] = RandomForestClassifier(max_samples=0.3)
models['40'] = RandomForestClassifier(max_samples=0.4)
models['50'] = RandomForestClassifier(max_samples=0.5)
models['60'] = RandomForestClassifier(max_samples=0.6)
models['70'] = RandomForestClassifier(max_samples=0.7)
models['80'] = RandomForestClassifier(max_samples=0.8)
models['90'] = RandomForestClassifier(max_samples=0.9)
models['100'] = RandomForestClassifier(max_samples=None)
return models
# evaluate a give model using cross-validation
def evaluate_model(model):
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
return scores
# define dataset
X, y = get_dataset()
# get the models to evaluate
models = get_models()
# evaluate the models and store results
results, names = list(), list()
for name, model in models.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print('>%s %.3f (%.3f)' % (name, mean(scores), std(scores)))
# plot model performance for comparison
pyplot.boxplot(results, labels=names, showmeans=True)
pyplot.xticks(rotation=45)
pyplot.show()
Running the example first reports the mean accuracy for each dataset size.
In this case, the results suggest that using a bootstrap sample size that is equal to the size of the training dataset achieves the best results on this dataset.
This is the default and it should probably be used in most cases.
>10 0.856 (0.031) >20 0.873 (0.029) >30 0.881 (0.021) >40 0.891 (0.033) >50 0.893 (0.025) >60 0.897 (0.030) >70 0.902 (0.024) >80 0.903 (0.024) >90 0.900 (0.026) >100 0.903 (0.027)
A box and whisker plot is created for the distribution of accuracy scores for each bootstrap sample size.
In this case, we can see a general trend that the larger the sample, the better the performance of the model.
You might like to extend this example and see what happens if the bootstrap sample size is larger or even much larger than the training dataset (e.g. you can set an integer value as the number of samples instead of a float percentage of the training dataset size).
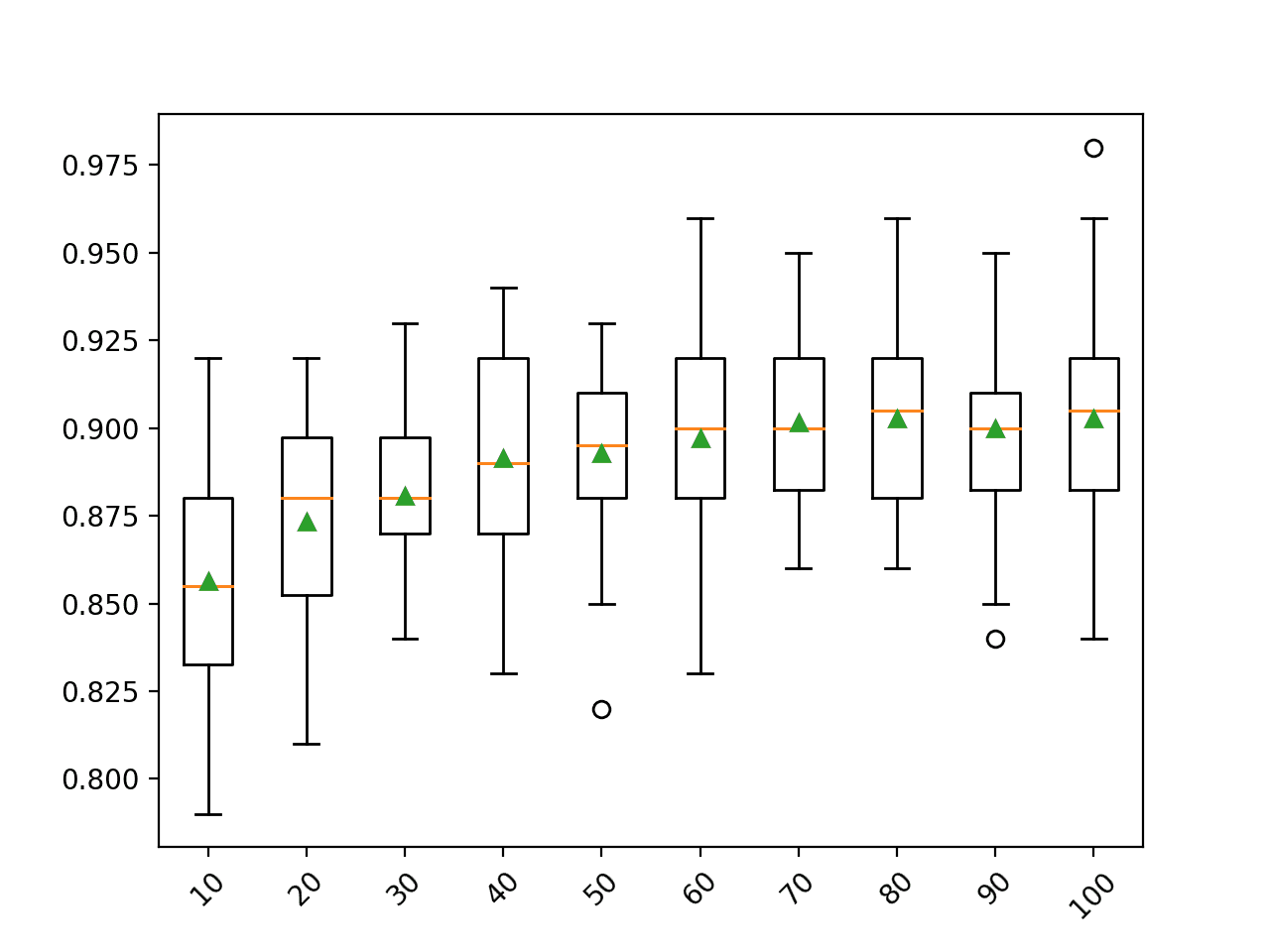
Box Plot of Random Forest Bootstrap Sample Size vs. Classification Accuracy
Explore Number of Features
The number of features that is randomly sampled for each split point is perhaps the most important feature to configure for random forest.
It is set via the max_features argument and defaults to the square root of the number of input features. In this case, for our test dataset, this would be sqrt(20) or about four features.
The example below explores the effect of the number of features randomly selected at each split point on model accuracy. We will try values from 1 to 7 and would expect a small value, around four, to perform well based on the heuristic.
# explore random forest number of features effect on performance
from numpy import mean
from numpy import std
from sklearn.datasets import make_classification
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.ensemble import RandomForestClassifier
from matplotlib import pyplot
# get the dataset
def get_dataset():
X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=3)
return X, y
# get a list of models to evaluate
def get_models():
models = dict()
models['1'] = RandomForestClassifier(max_features=1)
models['2'] = RandomForestClassifier(max_features=2)
models['3'] = RandomForestClassifier(max_features=3)
models['4'] = RandomForestClassifier(max_features=4)
models['5'] = RandomForestClassifier(max_features=5)
models['6'] = RandomForestClassifier(max_features=6)
models['7'] = RandomForestClassifier(max_features=7)
return models
# evaluate a give model using cross-validation
def evaluate_model(model):
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
return scores
# define dataset
X, y = get_dataset()
# get the models to evaluate
models = get_models()
# evaluate the models and store results
results, names = list(), list()
for name, model in models.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print('>%s %.3f (%.3f)' % (name, mean(scores), std(scores)))
# plot model performance for comparison
pyplot.boxplot(results, labels=names, showmeans=True)
pyplot.show()
Running the example first reports the mean accuracy for each feature set size.
In this case, the results suggest that a value between three and five would be appropriate, confirming the sensible default of four on this dataset. A value of five might even be better given the smaller standard deviation in classification accuracy as compared to a value of three or four.
>1 0.897 (0.023) >2 0.900 (0.028) >3 0.903 (0.027) >4 0.903 (0.022) >5 0.903 (0.019) >6 0.898 (0.025) >7 0.900 (0.024)
A box and whisker plot is created for the distribution of accuracy scores for each feature set size.
We can see a trend in performance rising and peaking with values between three and five and falling again as larger feature set sizes are considered.
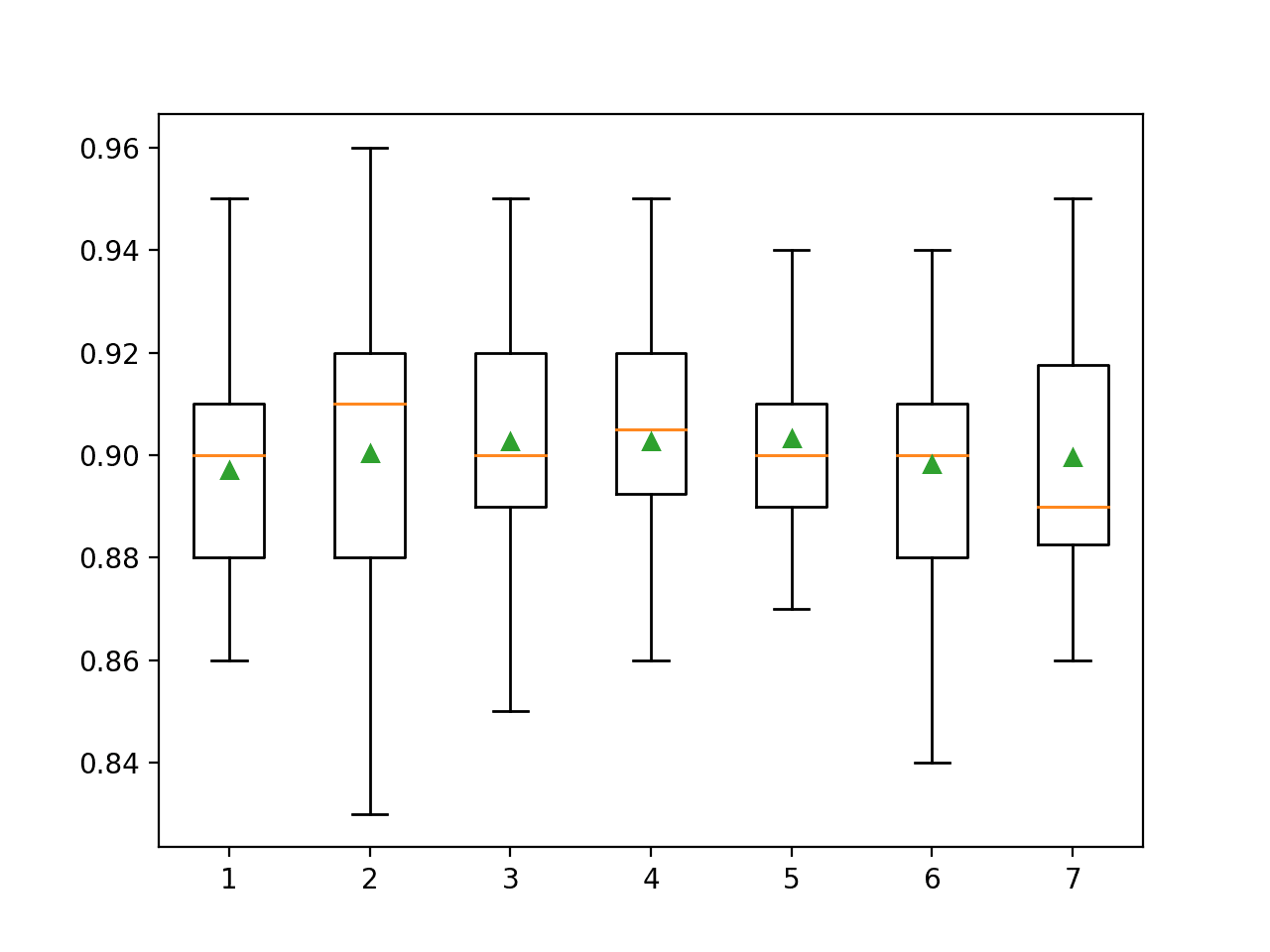
Box Plot of Random Forest Feature Set Size vs. Classification Accuracy
Explore Number of Trees
The number of trees is another key hyperparameter to configure for the random forest.
Typically, the number of trees is increased until the model performance stabilizes. Intuition might suggest that more trees will lead to overfitting, although this is not the case. Both bagging and random forest algorithms appear to be somewhat immune to overfitting the training dataset given the stochastic nature of the learning algorithm.
The number of trees can be set via the “n_estimators” argument and defaults to 100.
The example below explores the effect of the number of trees with values between 10 to 1,000.
# explore random forest number of trees effect on performance
from numpy import mean
from numpy import std
from sklearn.datasets import make_classification
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.ensemble import RandomForestClassifier
from matplotlib import pyplot
# get the dataset
def get_dataset():
X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=3)
return X, y
# get a list of models to evaluate
def get_models():
models = dict()
models['10'] = RandomForestClassifier(n_estimators=10)
models['50'] = RandomForestClassifier(n_estimators=50)
models['100'] = RandomForestClassifier(n_estimators=100)
models['500'] = RandomForestClassifier(n_estimators=500)
models['1000'] = RandomForestClassifier(n_estimators=1000)
return models
# evaluate a give model using cross-validation
def evaluate_model(model):
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
return scores
# define dataset
X, y = get_dataset()
# get the models to evaluate
models = get_models()
# evaluate the models and store results
results, names = list(), list()
for name, model in models.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print('>%s %.3f (%.3f)' % (name, mean(scores), std(scores)))
# plot model performance for comparison
pyplot.boxplot(results, labels=names, showmeans=True)
pyplot.show()
Running the example first reports the mean accuracy for each configured number of trees.
In this case, we can see that performance rises and stays flat after about 100 trees. Mean accuracy scores fluctuate across 100, 500, and 1,000 trees and this may be statistical noise.
>10 0.870 (0.036) >50 0.900 (0.028) >100 0.910 (0.024) >500 0.904 (0.024) >1000 0.906 (0.023)
A box and whisker plot is created for the distribution of accuracy scores for each configured number of trees.
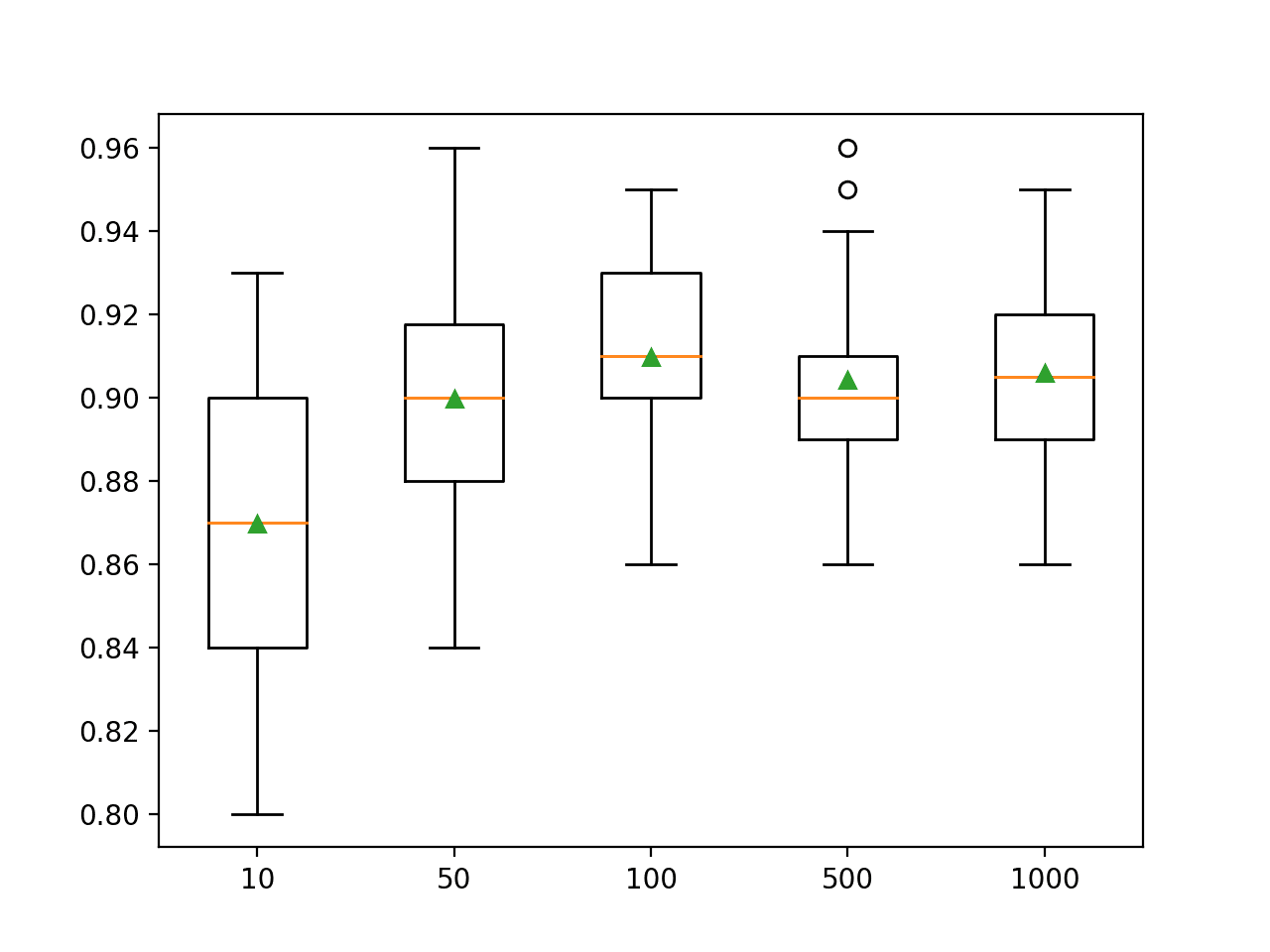
Box Plot of Random Forest Ensemble Size vs. Classification Accuracy
Explore Tree Depth
A final interesting hyperparameter is the maximum depth of decision trees used in the ensemble.
By default, trees are constructed to an arbitrary depth and are not pruned. This is a sensible default, although we can also explore fitting trees with different fixed depths.
The maximum tree depth can be specified via the max_depth argument and is set to None (no maximum depth) by default.
The example below explores the effect of random forest maximum tree depth on model performance.
# explore random forest tree depth effect on performance
from numpy import mean
from numpy import std
from sklearn.datasets import make_classification
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.ensemble import RandomForestClassifier
from matplotlib import pyplot
# get the dataset
def get_dataset():
X, y = make_classification(n_samples=1000, n_features=20, n_informative=15, n_redundant=5, random_state=3)
return X, y
# get a list of models to evaluate
def get_models():
models = dict()
models['1'] = RandomForestClassifier(max_depth=1)
models['2'] = RandomForestClassifier(max_depth=2)
models['3'] = RandomForestClassifier(max_depth=3)
models['4'] = RandomForestClassifier(max_depth=4)
models['5'] = RandomForestClassifier(max_depth=5)
models['6'] = RandomForestClassifier(max_depth=6)
models['7'] = RandomForestClassifier(max_depth=7)
models['None'] = RandomForestClassifier(max_depth=None)
return models
# evaluate a give model using cross-validation
def evaluate_model(model):
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
return scores
# define dataset
X, y = get_dataset()
# get the models to evaluate
models = get_models()
# evaluate the models and store results
results, names = list(), list()
for name, model in models.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print('>%s %.3f (%.3f)' % (name, mean(scores), std(scores)))
# plot model performance for comparison
pyplot.boxplot(results, labels=names, showmeans=True)
pyplot.show()
Running the example first reports the mean accuracy for each configured maximum tree depth.
In this case, we can see that larger depth results in better model performance, with the default of no maximum depth achieving the best performance on this dataset.
>1 0.771 (0.040) >2 0.807 (0.037) >3 0.834 (0.034) >4 0.857 (0.030) >5 0.872 (0.025) >6 0.887 (0.024) >7 0.890 (0.025) >None 0.903 (0.027)
A box and whisker plot is created for the distribution of accuracy scores for each configured maximum tree depth.
In this case, we can see a trend of improved performance with increase in tree depth, supporting the default of no maximum depth.
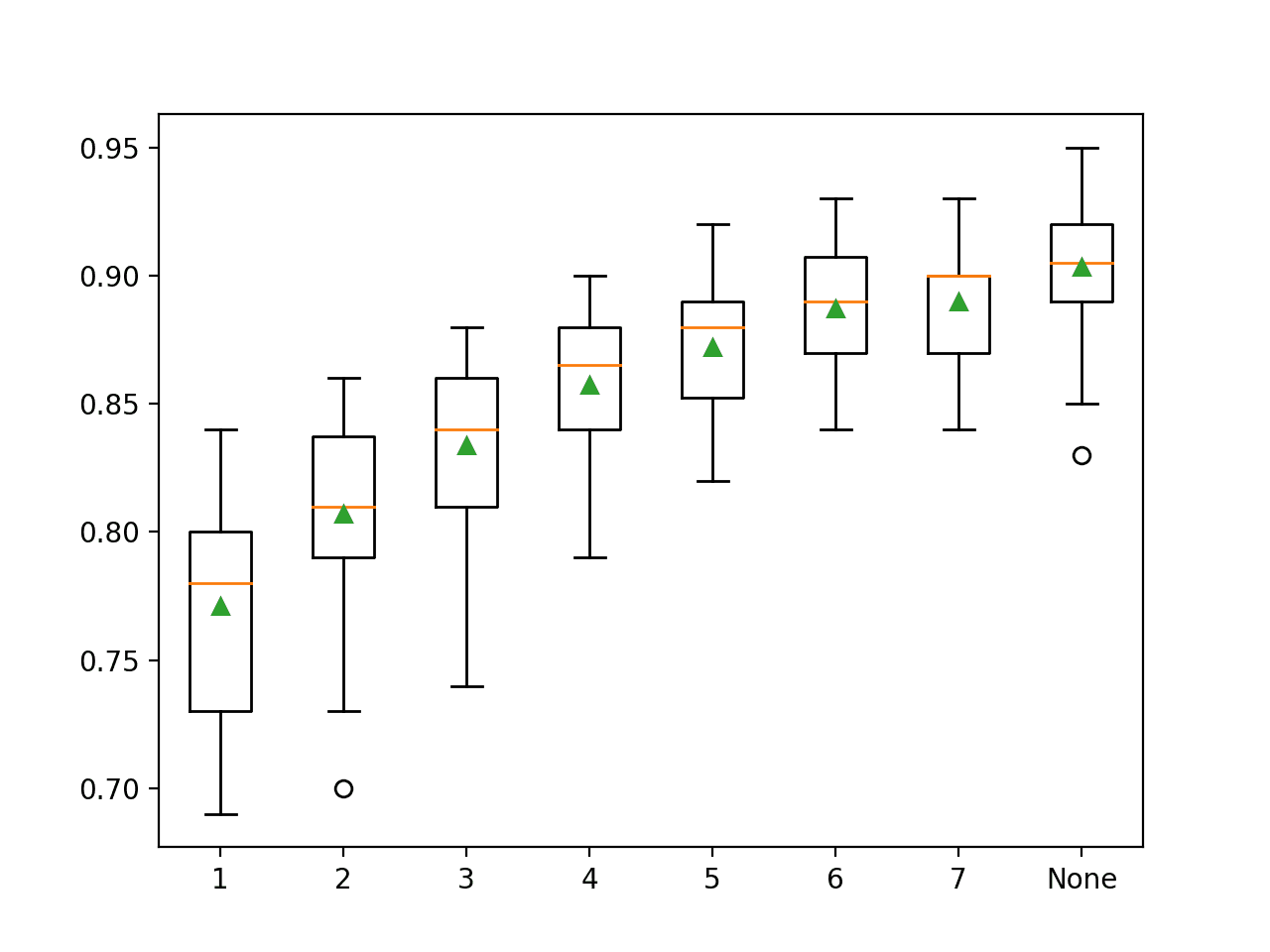
Box Plot of Random Forest Maximum Tree Depth vs. Classification Accuracy
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Tutorials
Books
- Applied Predictive Modeling, 2013.
- An Introduction to Statistical Learning with Applications in R, 2014.
APIs
Articles
Summary
In this tutorial, you discovered how to develop random forest ensembles for classification and regression.
Specifically, you learned:
- Random forest ensemble is an ensemble of decision trees and a natural extension of bagging.
- How to use the random forest ensemble for classification and regression with scikit-learn.
- How to explore the effect of random forest model hyperparameters on model performance.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.
The post How to Develop a Random Forest Ensemble in Python appeared first on Machine Learning Mastery.
