Author: Jason Brownlee
Model ensembles can achieve lower generalization error than single models but are challenging to develop with deep learning neural networks given the computational cost of training each single model.
An alternative is to train multiple model snapshots during a single training run and combine their predictions to make an ensemble prediction. A limitation of this approach is that the saved models will be similar, resulting in similar predictions and predictions errors and not offering much benefit from combining their predictions.
Effective ensembles require a diverse set of skillful ensemble members that have differing distributions of prediction errors. One approach to promoting a diversity of models saved during a single training run is to use an aggressive learning rate schedule that forces large changes in the model weights and, in turn, the nature of the model saved at each snapshot.
In this tutorial, you will discover how to develop snapshot ensembles of models saved using an aggressive learning rate schedule over a single training run.
After completing this tutorial, you will know:
- Snapshot ensembles combine the predictions from multiple models saved during a single training run.
- Diversity in model snapshots can be achieved through the use of aggressively cycling the learning rate used during a single training run.
- How to save model snapshots during a single run and load snapshot models to make ensemble predictions.
Let’s get started.

How to Develop a Snapshot Ensemble Deep Learning Neural Network in Python With Keras
Photo by Jason Jacobs, some rights reserved.
Tutorial Overview
This tutorial is divided into five parts; they are:
- Snapshot Ensembles
- Multi-Class Classification Problem
- Multilayer Perceptron Model
- Cosine Annealing Learning Rate
- MLP Snapshot Ensemble
Snapshot Ensembles
A problem with ensemble learning with deep learning methods is the large computational cost of training multiple models.
This is because of the use of very deep models and very large datasets that can result in model training times that may extend to days, weeks, or even months.
Despite its obvious advantages, the use of ensembling for deep networks is not nearly as widespread as it is for other algorithms. One likely reason for this lack of adaptation may be the cost of learning multiple neural networks. Training deep networks can last for weeks, even on high performance hardware with GPU acceleration.
— Snapshot Ensembles: Train 1, get M for free, 2017.
One approach to ensemble learning for deep learning neural networks is to collect multiple models from a single training run. This addresses the computational cost of training multiple deep learning models as models can be selected and saved during training, then used to make an ensemble prediction.
A key benefit of ensemble learning is in improved performance compared to the predictions from single models. This can be achieved through the selection of members that have good skill, but in different ways, providing a diverse set of predictions to be combined. A limitation of collecting multiple models during a single training run is that the models may be good, but too similar.
This can be addressed by changing the learning algorithm for the deep neural network to force the exploration of different network weights during a single training run that will result, in turn, with models that have differing performance. One way that this can be achieved is by aggressively changing the learning rate used during training.
An approach to systematically and aggressively changing the learning rate during training to result in very different network weights is referred to as “Stochastic Gradient Descent with Warm Restarts” or SGDR for short, described by Ilya Loshchilov and Frank Hutter in their 2017 paper “SGDR: Stochastic Gradient Descent with Warm Restarts.”
Their approach involves systematically changing the learning rate over training epochs, called cosine annealing. This approach requires the specification of two hyperparameters: the initial learning rate and the total number of training epochs.
The “cosine annealing” method has the effect of starting with a large learning rate that is relatively rapidly decreased to a minimum value before being dramatically increased again. The model weights are subjected to the dramatic changes during training, having the effect of using “good weights” as the starting point for the subsequent learning rate cycle, but allowing the learning algorithm to converge to a different solution.
The resetting of the learning rate acts like a simulated restart of the learning process and the re-use of good weights as the starting point of the restart is referred to as a “warm restart,” in contrast to a “cold restart” where a new set of small random numbers may be used as a starting point.
The “good weights” at the bottom of each cycle can be saved to file, providing a snapshot of the model. These snapshots can be collected together at the end of the run and used in a model averaging ensemble. The saving and use of these models during an aggressive learning rate schedule is referred to as a “Snapshot Ensemble” and was described by Gao Huang, et al. in their 2017 paper titled “Snapshot Ensembles: Train 1, get M for free” and subsequently also used in an updated version of the Loshchilov and Hutter paper.
… we let SGD converge M times to local minima along its optimization path. Each time the model converges, we save the weights and add the corresponding network to our ensemble. We then restart the optimization with a large learning rate to escape the current local minimum.
— Snapshot Ensembles: Train 1, get M for free, 2017.
The ensemble of models is created during the course of training a single model, therefore, the authors claim that the ensemble forecast is provided at no additional cost.
[the approach allows] learning an ensemble of multiple neural networks without incurring any additional training costs.
— Snapshot Ensembles: Train 1, get M for free, 2017.
Although a cosine annealing schedule is used for the learning rate, other aggressive learning rate schedules could be used, such as the simpler cyclical learning rate schedule described by Leslie Smith in the 2017 paper titled “Cyclical Learning Rates for Training Neural Networks.”
Now that we are familiar with the snapshot ensemble technique, we can look at how to implement it in Python with Keras.
Want Better Results with Deep Learning?
Take my free 7-day email crash course now (with sample code).
Click to sign-up and also get a free PDF Ebook version of the course.
Multi-Class Classification Problem
We will use a small multi-class classification problem as the basis to demonstrate the snapshot ensemble.
The scikit-learn class provides the make_blobs() function that can be used to create a multi-class classification problem with the prescribed number of samples, input variables, classes, and variance of samples within a class.
The problem has two input variables (to represent the x and y coordinates of the points) and a standard deviation of 2.0 for points within each group. We will use the same random state (seed for the pseudorandom number generator) to ensure that we always get the same data points.
# generate 2d classification dataset X, y = make_blobs(n_samples=1000, centers=3, n_features=2, cluster_std=2, random_state=2)
The result is the input and output elements of a dataset that we can model.
In order to get a feeling for the complexity of the problem, we can plot each point on a two-dimensional scatter plot and color each point by class value.
The complete example is listed below.
# scatter plot of blobs dataset from sklearn.datasets.samples_generator import make_blobs from matplotlib import pyplot from numpy import where # generate 2d classification dataset X, y = make_blobs(n_samples=1000, centers=3, n_features=2, cluster_std=2, random_state=2) # scatter plot for each class value for class_value in range(3): # select indices of points with the class label row_ix = where(y == class_value) # scatter plot for points with a different color pyplot.scatter(X[row_ix, 0], X[row_ix, 1]) # show plot pyplot.show()
Running the example creates a scatter plot of the entire dataset. We can see that the standard deviation of 2.0 means that the classes are not linearly separable (separable by a line) causing many ambiguous points.
This is desirable as it means that the problem is non-trivial and will allow a neural network model to find many different “good enough” candidate solutions resulting in a high variance.
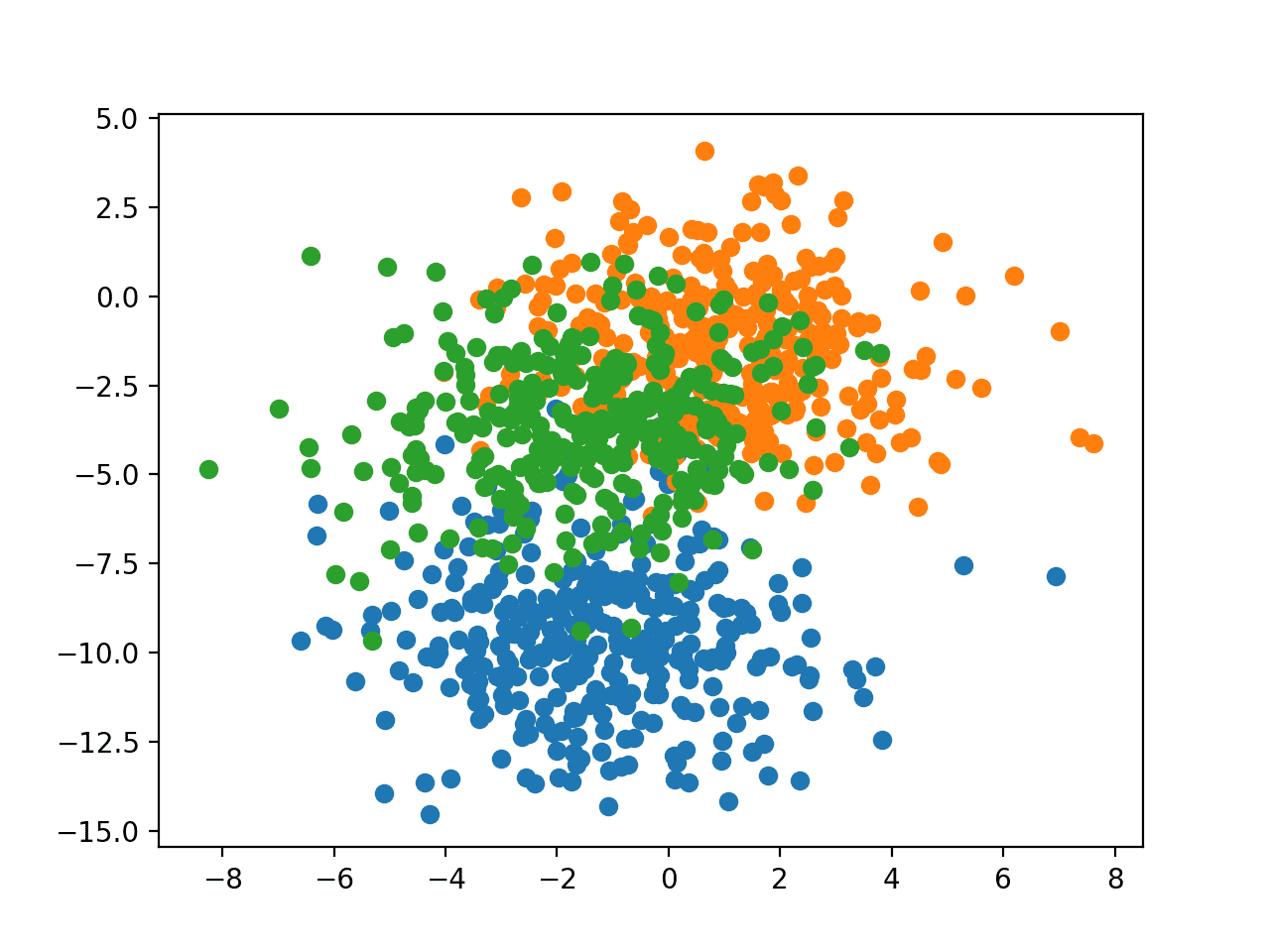
Scatter Plot of Blobs Dataset With Three Classes and Points Colored by Class Value
Multilayer Perceptron Model
Before we define a model, we need to contrive a problem that is appropriate for the ensemble.
In our problem, the training dataset is relatively small. Specifically, there is a 10:1 ratio of examples in the training dataset to the holdout dataset. This mimics a situation where we may have a vast number of unlabeled examples and a small number of labeled examples with which to train a model.
We will create 1,100 data points from the blobs problem. The model will be trained on the first 100 points and the remaining 1,000 will be held back in a test dataset, unavailable to the model.
The problem is a multi-class classification problem, and we will model it using a softmax activation function on the output layer. This means that the model will predict a vector with three elements with the probability that the sample belongs to each of the three classes. Therefore, we must one hot encode the class values before we split the rows into the train and test datasets. We can do this using the Keras to_categorical() function.
# generate 2d classification dataset X, y = make_blobs(n_samples=1100, centers=3, n_features=2, cluster_std=2, random_state=2) # one hot encode output variable y = to_categorical(y) # split into train and test n_train = 100 trainX, testX = X[:n_train, :], X[n_train:, :] trainy, testy = y[:n_train], y[n_train:]
Next, we can define and compile the model.
The model will expect samples with two input variables. The model then has a single hidden layer with 25 nodes and a rectified linear activation function, then an output layer with three nodes to predict the probability of each of the three classes and a softmax activation function.
Because the problem is multi-class, we will use the categorical cross entropy loss function to optimize the model and stochastic gradient descent with a small learning rate and momentum.
# define model model = Sequential() model.add(Dense(25, input_dim=2, activation='relu')) model.add(Dense(3, activation='softmax')) opt = SGD(lr=0.01, momentum=0.9) model.compile(loss='categorical_crossentropy', optimizer=opt, metrics=['accuracy'])
The model is fit for 200 training epochs and we will evaluate the model each epoch on the test set, using the test set as a validation set.
# fit model history = model.fit(trainX, trainy, validation_data=(testX, testy), epochs=200, verbose=0)
At the end of the run, we will evaluate the performance of the model on the train and test sets.
# evaluate the model
_, train_acc = model.evaluate(trainX, trainy, verbose=0)
_, test_acc = model.evaluate(testX, testy, verbose=0)
print('Train: %.3f, Test: %.3f' % (train_acc, test_acc))
Then finally, we will plot learning curves of the model accuracy over each training epoch on both the training and validation datasets.
# learning curves of model accuracy pyplot.plot(history.history['acc'], label='train') pyplot.plot(history.history['val_acc'], label='test') pyplot.legend() pyplot.show()
Tying all of this together, the complete example is listed below.
# develop an mlp for blobs dataset
from sklearn.datasets.samples_generator import make_blobs
from keras.utils import to_categorical
from keras.models import Sequential
from keras.layers import Dense
from keras.optimizers import SGD
from matplotlib import pyplot
# generate 2d classification dataset
X, y = make_blobs(n_samples=1100, centers=3, n_features=2, cluster_std=2, random_state=2)
# one hot encode output variable
y = to_categorical(y)
# split into train and test
n_train = 100
trainX, testX = X[:n_train, :], X[n_train:, :]
trainy, testy = y[:n_train], y[n_train:]
# define model
model = Sequential()
model.add(Dense(25, input_dim=2, activation='relu'))
model.add(Dense(3, activation='softmax'))
opt = SGD(lr=0.01, momentum=0.9)
model.compile(loss='categorical_crossentropy', optimizer=opt, metrics=['accuracy'])
# fit model
history = model.fit(trainX, trainy, validation_data=(testX, testy), epochs=200, verbose=0)
# evaluate the model
_, train_acc = model.evaluate(trainX, trainy, verbose=0)
_, test_acc = model.evaluate(testX, testy, verbose=0)
print('Train: %.3f, Test: %.3f' % (train_acc, test_acc))
# learning curves of model accuracy
pyplot.plot(history.history['acc'], label='train')
pyplot.plot(history.history['val_acc'], label='test')
pyplot.legend()
pyplot.show()
Running the example prints the performance of the final model on the train and test datasets.
Your specific results will vary (by design!) given the high variance nature of the model.
In this case, we can see that the model achieved about 84% accuracy on the training dataset, which we know is optimistic, and about 79% on the test dataset, which we would expect to be more realistic.
Train: 0.840, Test: 0.796
A line plot is also created showing the learning curves for the model accuracy on the train and test sets over each training epoch.
We can see that training accuracy is more optimistic over most of the run as we also noted with the final scores.
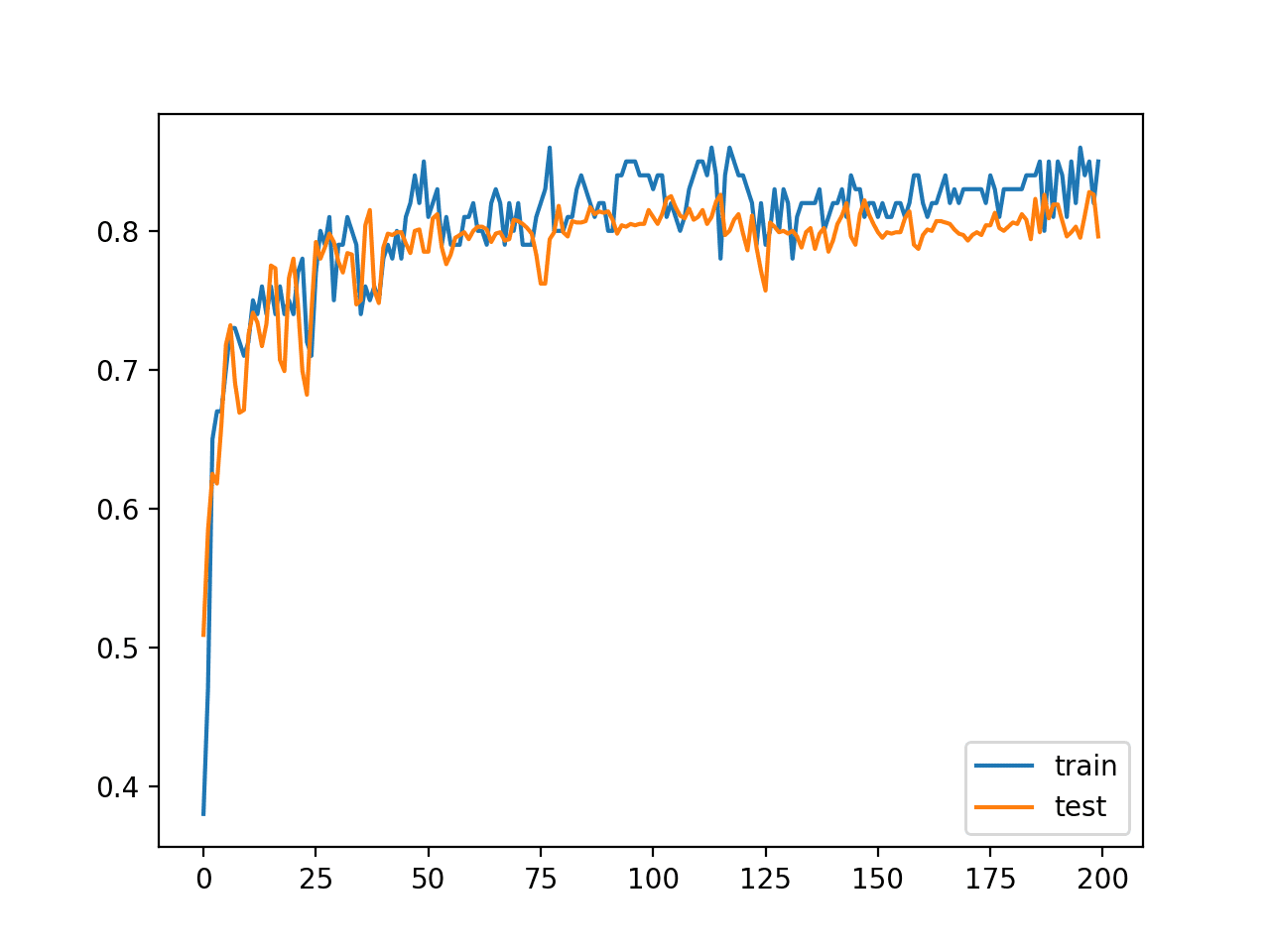
Line Plot Learning Curves of Model Accuracy on Train and Test Dataset over Each Training Epoch
Next, we can look at how to implement an aggressive learning rate schedule.
Cosine Annealing Learning Rate
An effective snapshot ensemble requires training a neural network with an aggressive learning rate schedule.
The cosine annealing schedule is an example of an aggressive learning rate schedule where learning rate starts high and is dropped relatively rapidly to a minimum value near zero before being increased again to the maximum.
We can implement the schedule as described in the 2017 paper “Snapshot Ensembles: Train 1, get M for free.” The equation requires the total training epochs, maximum learning rate, and number of cycles as arguments as well as the current epoch number. The function then returns the learning rate for the given epoch.
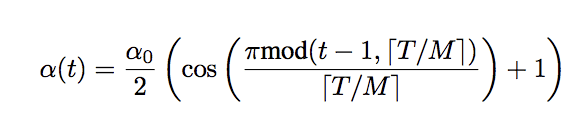
Equation for the Cosine Annealing Learning Rate Schedule
Where a(t) is the learning rate at epoch t, a0 is the maximum learning rate, T is the total epochs, M is the number of cycles, mod is the modulo operation, and square brackets indicate a floor operation.
Taken from “Snapshot Ensembles: Train 1, get M for free”.
The function cosine_annealing() below implements the equation.
# cosine annealing learning rate schedule def cosine_annealing(epoch, n_epochs, n_cycles, lrate_max): epochs_per_cycle = floor(n_epochs/n_cycles) cos_inner = (pi * (epoch % epochs_per_cycle)) / (epochs_per_cycle) return lrate_max/2 * (cos(cos_inner) + 1)
We can test this implementation by plotting the learning rate over 100 epochs with five cycles (e.g. 20 epochs long) and a maximum learning rate of 0.01. The complete example is listed below.
# example of a cosine annealing learning rate schedule from matplotlib import pyplot from math import pi from math import cos from math import floor # cosine annealing learning rate schedule def cosine_annealing(epoch, n_epochs, n_cycles, lrate_max): epochs_per_cycle = floor(n_epochs/n_cycles) cos_inner = (pi * (epoch % epochs_per_cycle)) / (epochs_per_cycle) return lrate_max/2 * (cos(cos_inner) + 1) # create learning rate series n_epochs = 100 n_cycles = 5 lrate_max = 0.01 series = [cosine_annealing(i, n_epochs, n_cycles, lrate_max) for i in range(n_epochs)] # plot series pyplot.plot(series) pyplot.show()
Running the example creates a line plot of the learning rate schedule over 100 epochs.
We can see that the learning rate starts at the maximum value at epoch 0 and decreases rapidly to epoch 19, before being reset at epoch 20, the start of the next cycle. The cycle is repeated five times as specified in the argument.
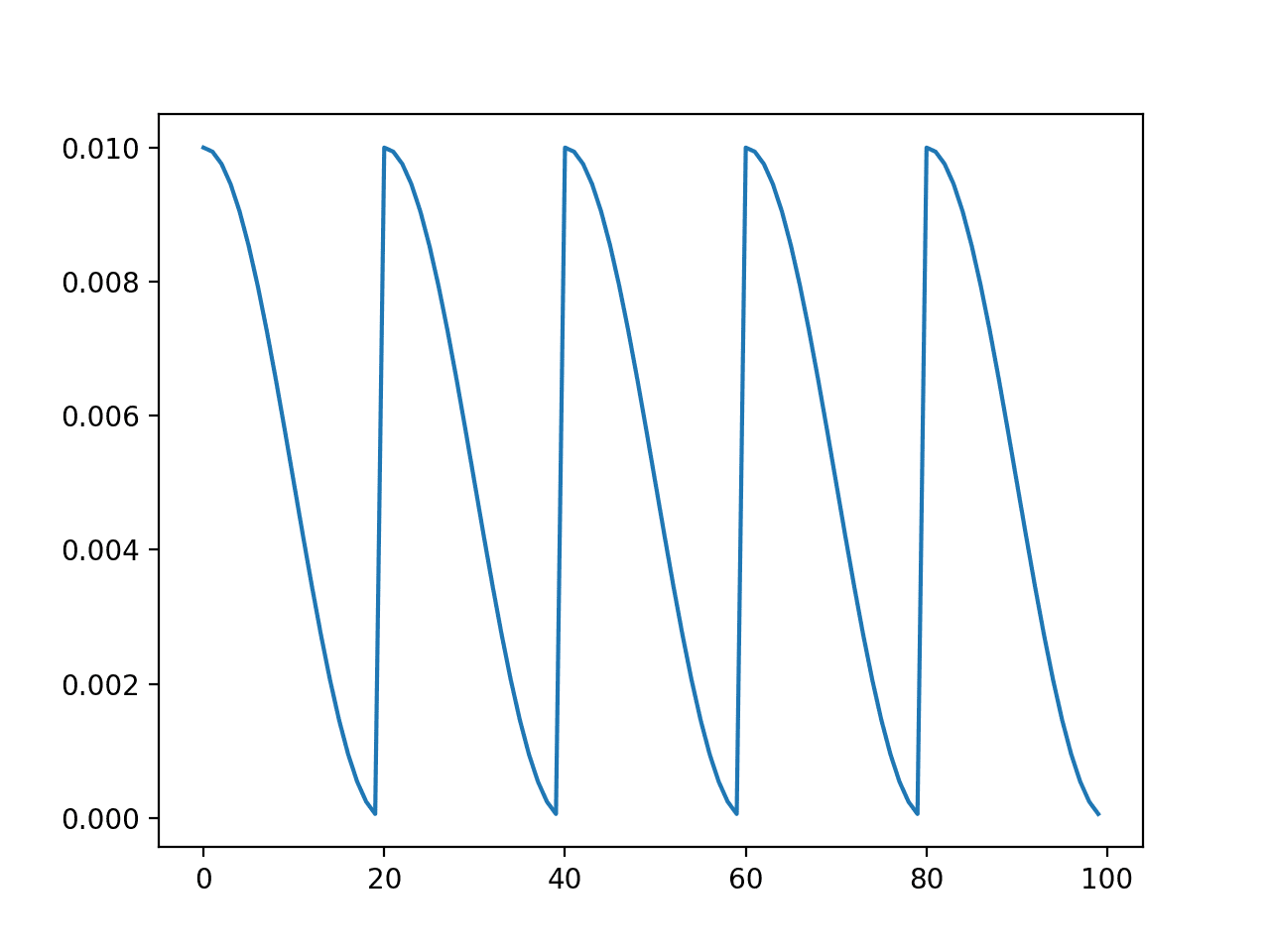
Line Plot of Cosine Annealing Learning Rate Schedule
We can implement this schedule as a custom callback in Keras. This allows the parameters of the schedule to be specified and for the learning rate to be logged so we can ensure it had the desired effect.
A custom callback can be defined as a Python class that extends the Keras Callback class.
In the class constructor, we can take the required configuration as arguments and save them for use, specifically the total number of training epochs, the number of cycles for the learning rate schedule, and the maximum learning rate.
We can use our cosine_annealing() defined above to calculate the learning rate for a given training epoch.
The Callback class allows an on_epoch_begin() function to be overridden that will be called prior to each training epoch. We can override this function to calculate the learning rate for the current epoch and set it in the optimizer. We can also keep track of the learning rate in an internal list.
The complete custom callback is defined below.
# define custom learning rate schedule class CosineAnnealingLearningRateSchedule(Callback): # constructor def __init__(self, n_epochs, n_cycles, lrate_max, verbose=0): self.epochs = n_epochs self.cycles = n_cycles self.lr_max = lrate_max self.lrates = list() # calculate learning rate for an epoch def cosine_annealing(self, epoch, n_epochs, n_cycles, lrate_max): epochs_per_cycle = floor(n_epochs/n_cycles) cos_inner = (pi * (epoch % epochs_per_cycle)) / (epochs_per_cycle) return lrate_max/2 * (cos(cos_inner) + 1) # calculate and set learning rate at the start of the epoch def on_epoch_begin(self, epoch, logs=None): # calculate learning rate lr = self.cosine_annealing(epoch, self.epochs, self.cycles, self.lr_max) # set learning rate backend.set_value(self.model.optimizer.lr, lr) # log value self.lrates.append(lr)
We can create an instance of the callback and set the arguments. We will train the model for 400 epochs and set the number of cycles to be 50 epochs long, or 500 / 50, a suggestion made and configuration used throughout the snapshot ensembles paper.
We lower the learning rate at a very fast pace, encouraging the model to converge towards its first local minimum after as few as 50 epochs.
— Snapshot Ensembles: Train 1, get M for free, 2017.
The paper also suggests that the learning rate can be set each sample or each mini-batch instead of prior to each epoch to give more nuance to the updates, but we will leave this as a future exercise.
… we update the learning rate at each iteration rather than at every epoch. This improves the convergence of short cycles, even when a large initial learning rate is used.
— Snapshot Ensembles: Train 1, get M for free, 2017.
Once the callback is instantiated and configured, we can specify it as part of the list of callbacks to the call to the fit() function to train the model.
# define learning rate callback n_epochs = 400 n_cycles = n_epochs / 50 ca = CosineAnnealingLearningRateSchedule(n_epochs, n_cycles, 0.01) # fit model history = model.fit(trainX, trainy, validation_data=(testX, testy), epochs=n_epochs, verbose=0, callbacks=[ca])
At the end of the run, we can confirm that the learning rate schedule was performed by plotting the contents of the lrates list.
# plot learning rate pyplot.plot(ca.lrates) pyplot.show()
Tying these elements together, the complete example of training an MLP on the blobs problem with a cosine annealing learning rate schedule is listed below.
# mlp with cosine annealing learning rate schedule on blobs problem
from sklearn.datasets.samples_generator import make_blobs
from keras.utils import to_categorical
from keras.models import Sequential
from keras.layers import Dense
from keras.callbacks import Callback
from keras.optimizers import SGD
from keras import backend
from math import pi
from math import cos
from math import floor
from matplotlib import pyplot
# define custom learning rate schedule
class CosineAnnealingLearningRateSchedule(Callback):
# constructor
def __init__(self, n_epochs, n_cycles, lrate_max, verbose=0):
self.epochs = n_epochs
self.cycles = n_cycles
self.lr_max = lrate_max
self.lrates = list()
# calculate learning rate for an epoch
def cosine_annealing(self, epoch, n_epochs, n_cycles, lrate_max):
epochs_per_cycle = floor(n_epochs/n_cycles)
cos_inner = (pi * (epoch % epochs_per_cycle)) / (epochs_per_cycle)
return lrate_max/2 * (cos(cos_inner) + 1)
# calculate and set learning rate at the start of the epoch
def on_epoch_begin(self, epoch, logs=None):
# calculate learning rate
lr = self.cosine_annealing(epoch, self.epochs, self.cycles, self.lr_max)
# set learning rate
backend.set_value(self.model.optimizer.lr, lr)
# log value
self.lrates.append(lr)
# generate 2d classification dataset
X, y = make_blobs(n_samples=1100, centers=3, n_features=2, cluster_std=2, random_state=2)
# one hot encode output variable
y = to_categorical(y)
# split into train and test
n_train = 100
trainX, testX = X[:n_train, :], X[n_train:, :]
trainy, testy = y[:n_train], y[n_train:]
# define model
model = Sequential()
model.add(Dense(25, input_dim=2, activation='relu'))
model.add(Dense(3, activation='softmax'))
opt = SGD(momentum=0.9)
model.compile(loss='categorical_crossentropy', optimizer=opt, metrics=['accuracy'])
# define learning rate callback
n_epochs = 400
n_cycles = n_epochs / 50
ca = CosineAnnealingLearningRateSchedule(n_epochs, n_cycles, 0.01)
# fit model
history = model.fit(trainX, trainy, validation_data=(testX, testy), epochs=n_epochs, verbose=0, callbacks=[ca])
# evaluate the model
_, train_acc = model.evaluate(trainX, trainy, verbose=0)
_, test_acc = model.evaluate(testX, testy, verbose=0)
print('Train: %.3f, Test: %.3f' % (train_acc, test_acc))
# plot learning rate
pyplot.plot(ca.lrates)
pyplot.show()
# learning curves of model accuracy
pyplot.plot(history.history['acc'], label='train')
pyplot.plot(history.history['val_acc'], label='test')
pyplot.legend()
pyplot.show()
Running the example first reports the accuracy of the model on the training and test sets.
Your results will vary given the stochastic nature of the training algorithm.
In this case, we do not see much difference in the performance of the final model as compared to the previous section.
Train: 0.850, Test: 0.806
A line plot of the learning rate schedule is created, showing eight cycles of 50 epochs each.

Cosine Annealing Learning Rate Schedule While Fitting an MLP on the Blobs Problem
Finally, a line plot of model accuracy on the train and test sets is created over each training epoch.
We can see that although the learning rate was changed dramatically, there was not a dramatic effect on model accuracy, likely because the chosen classification problem is not very difficult.

Line Plot of Train and Test Set Accuracy on the Blobs Dataset With a Cosine Annealing Learning Rate Schedule
Now that we know how to implement the cosine annealing learning schedule, we can use it to prepare a snapshot ensemble.
MLP Snapshot Ensemble
We can develop a snapshot ensemble in two parts.
The first part involves creating a custom callback to save the model at the bottom of each learning rate schedule. The second part involves loading the saved models and using them to make an ensemble prediction.
Save Snapshot Models During Training
The CosineAnnealingLearningRateSchedule can be updated to override the on_epoch_end() function called at the end of each training epoch. In this function, we can check if the current epoch that just ended was the end of a cycle. If so, we can save the model to file.
Below is the updated callback, named the SnapshotEnsemble class.
A debug message is printed each time a model is saved as confirmation that models are being saved at the right time. For example, with 50-epoch long cycles, we would expect a model to be saved on epoch 49, 99, etc. and the learning rate reset at epoch 50, 100, etc.
# snapshot ensemble with custom learning rate schedule
class SnapshotEnsemble(Callback):
# constructor
def __init__(self, n_epochs, n_cycles, lrate_max, verbose=0):
self.epochs = n_epochs
self.cycles = n_cycles
self.lr_max = lrate_max
self.lrates = list()
# calculate learning rate for epoch
def cosine_annealing(self, epoch, n_epochs, n_cycles, lrate_max):
epochs_per_cycle = floor(n_epochs/n_cycles)
cos_inner = (pi * (epoch % epochs_per_cycle)) / (epochs_per_cycle)
return lrate_max/2 * (cos(cos_inner) + 1)
# calculate and set learning rate at the start of the epoch
def on_epoch_begin(self, epoch, logs={}):
# calculate learning rate
lr = self.cosine_annealing(epoch, self.epochs, self.cycles, self.lr_max)
# set learning rate
backend.set_value(self.model.optimizer.lr, lr)
# log value
self.lrates.append(lr)
# save models at the end of each cycle
def on_epoch_end(self, epoch, logs={}):
# check if we can save model
epochs_per_cycle = floor(self.epochs / self.cycles)
if epoch != 0 and (epoch + 1) % epochs_per_cycle == 0:
# save model to file
filename = "snapshot_model_%d.h5" % int((epoch + 1) / epochs_per_cycle)
self.model.save(filename)
print('>saved snapshot %s, epoch %d' % (filename, epoch))
We will train the model for 500 epochs, to give 10 models to choose from later when making an ensemble prediction.
The complete example of using this new snapshot ensemble to save models to file is listed below.
# example of saving models for a snapshot ensemble
from sklearn.datasets.samples_generator import make_blobs
from keras.utils import to_categorical
from keras.models import Sequential
from keras.layers import Dense
from keras.callbacks import Callback
from keras.optimizers import SGD
from keras import backend
from math import pi
from math import cos
from math import floor
# snapshot ensemble with custom learning rate schedule
class SnapshotEnsemble(Callback):
# constructor
def __init__(self, n_epochs, n_cycles, lrate_max, verbose=0):
self.epochs = n_epochs
self.cycles = n_cycles
self.lr_max = lrate_max
self.lrates = list()
# calculate learning rate for epoch
def cosine_annealing(self, epoch, n_epochs, n_cycles, lrate_max):
epochs_per_cycle = floor(n_epochs/n_cycles)
cos_inner = (pi * (epoch % epochs_per_cycle)) / (epochs_per_cycle)
return lrate_max/2 * (cos(cos_inner) + 1)
# calculate and set learning rate at the start of the epoch
def on_epoch_begin(self, epoch, logs={}):
# calculate learning rate
lr = self.cosine_annealing(epoch, self.epochs, self.cycles, self.lr_max)
# set learning rate
backend.set_value(self.model.optimizer.lr, lr)
# log value
self.lrates.append(lr)
# save models at the end of each cycle
def on_epoch_end(self, epoch, logs={}):
# check if we can save model
epochs_per_cycle = floor(self.epochs / self.cycles)
if epoch != 0 and (epoch + 1) % epochs_per_cycle == 0:
# save model to file
filename = "snapshot_model_%d.h5" % int((epoch + 1) / epochs_per_cycle)
self.model.save(filename)
print('>saved snapshot %s, epoch %d' % (filename, epoch))
# generate 2d classification dataset
X, y = make_blobs(n_samples=1100, centers=3, n_features=2, cluster_std=2, random_state=2)
# one hot encode output variable
y = to_categorical(y)
# split into train and test
n_train = 100
trainX, testX = X[:n_train, :], X[n_train:, :]
trainy, testy = y[:n_train], y[n_train:]
# define model
model = Sequential()
model.add(Dense(50, input_dim=2, activation='relu'))
model.add(Dense(3, activation='softmax'))
opt = SGD(momentum=0.9)
model.compile(loss='categorical_crossentropy', optimizer=opt, metrics=['accuracy'])
# create snapshot ensemble callback
n_epochs = 500
n_cycles = n_epochs / 50
ca = SnapshotEnsemble(n_epochs, n_cycles, 0.01)
# fit model
model.fit(trainX, trainy, validation_data=(testX, testy), epochs=n_epochs, verbose=0, callbacks=[ca])
Running the example reports that 10 models were saved for the 10-ends of the cosine annealing learning rate schedule.
>saved snapshot snapshot_model_1.h5, epoch 49 >saved snapshot snapshot_model_2.h5, epoch 99 >saved snapshot snapshot_model_3.h5, epoch 149 >saved snapshot snapshot_model_4.h5, epoch 199 >saved snapshot snapshot_model_5.h5, epoch 249 >saved snapshot snapshot_model_6.h5, epoch 299 >saved snapshot snapshot_model_7.h5, epoch 349 >saved snapshot snapshot_model_8.h5, epoch 399 >saved snapshot snapshot_model_9.h5, epoch 449 >saved snapshot snapshot_model_10.h5, epoch 499
Load Models and Make Ensemble Prediction
Once the snapshot models have been saved to file, they can be loaded and used to make an ensemble prediction.
The first step is to load the models into memory. For large models, this could be done one model at a time, make a prediction, and move on to the next model before combining predictions. In this case, the models are relatively small and we can load all 10 from file as a list.
# load models from file
def load_all_models(n_models):
all_models = list()
for i in range(n_models):
# define filename for this ensemble
filename = 'snapshot_model_' + str(i + 1) + '.h5'
# load model from file
model = load_model(filename)
# add to list of members
all_models.append(model)
print('>loaded %s' % filename)
return all_models
We would expect that models saved towards the end of the run may have better performance than models saved earlier in the run. As such, we can reverse the list of loaded models so that the older models are first.
# reverse loaded models so we build the ensemble with the last models first members = list(reversed(members))
We don’t know how many snapshots are required to make a good prediction for this problem. We can explore the effect of the number of ensemble members on test set accuracy by creating ensembles of increasing size starting with the final model at epoch 499, then adding the model saved at epoch 449, and so on until all 10 models are included.
First, we require a function to make a prediction given a list of models. Given that each model predicts the probabilities of each of the output classes, we can sum the predicted probabilities across the models and select the class with the most support via the argmax() function. The ensemble_predictions() function below implements this functionality.
# make an ensemble prediction for multi-class classification def ensemble_predictions(members, testX): # make predictions yhats = [model.predict(testX) for model in members] yhats = array(yhats) # sum across ensemble members summed = numpy.sum(yhats, axis=0) # argmax across classes result = argmax(summed, axis=1) return result
We can then evaluate an ensemble of a given size by selecting the first n members from the list of models, making a prediction by calling the ensemble_predictions() function, and then calculating and returning the accuracy of the prediction. The evaluate_n_members() function below implements this behavior.
# evaluate a specific number of members in an ensemble def evaluate_n_members(members, n_members, testX, testy): # select a subset of members subset = members[:n_members] # make prediction yhat = ensemble_predictions(subset, testX) # calculate accuracy return accuracy_score(testy, yhat)
The performance of each ensemble can also be contrasted with the performance of each standalone model and the average performance of all standalone models.
# evaluate different numbers of ensembles on hold out set
single_scores, ensemble_scores = list(), list()
for i in range(1, len(members)+1):
# evaluate model with i members
ensemble_score = evaluate_n_members(members, i, testX, testy)
# evaluate the i'th model standalone
testy_enc = to_categorical(testy)
_, single_score = members[i-1].evaluate(testX, testy_enc, verbose=0)
# summarize this step
print('> %d: single=%.3f, ensemble=%.3f' % (i, single_score, ensemble_score))
ensemble_scores.append(ensemble_score)
single_scores.append(single_score)
# summarize average accuracy of a single final model
print('Accuracy %.3f (%.3f)' % (mean(single_scores), std(single_scores)))
Finally, we can plot the performance of each individual snapshot model (blue dots) compared to the performance of an ensemble that includes all models up to and including each individual model (orange line).
# plot score vs number of ensemble members x_axis = [i for i in range(1, len(members)+1)] pyplot.plot(x_axis, single_scores, marker='o', linestyle='None') pyplot.plot(x_axis, ensemble_scores, marker='o') pyplot.show()
The complete example of making snapshot ensemble predictions with different sized ensembles is listed below.
# load models and make a snapshot ensemble prediction
from sklearn.datasets.samples_generator import make_blobs
from sklearn.metrics import accuracy_score
from keras.utils import to_categorical
from keras.models import load_model
from keras.models import Sequential
from keras.layers import Dense
from matplotlib import pyplot
from numpy import mean
from numpy import std
import numpy
from numpy import array
from numpy import argmax
# load models from file
def load_all_models(n_models):
all_models = list()
for i in range(n_models):
# define filename for this ensemble
filename = 'snapshot_model_' + str(i + 1) + '.h5'
# load model from file
model = load_model(filename)
# add to list of members
all_models.append(model)
print('>loaded %s' % filename)
return all_models
# make an ensemble prediction for multi-class classification
def ensemble_predictions(members, testX):
# make predictions
yhats = [model.predict(testX) for model in members]
yhats = array(yhats)
# sum across ensemble members
summed = numpy.sum(yhats, axis=0)
# argmax across classes
result = argmax(summed, axis=1)
return result
# evaluate a specific number of members in an ensemble
def evaluate_n_members(members, n_members, testX, testy):
# select a subset of members
subset = members[:n_members]
# make prediction
yhat = ensemble_predictions(subset, testX)
# calculate accuracy
return accuracy_score(testy, yhat)
# generate 2d classification dataset
X, y = make_blobs(n_samples=1100, centers=3, n_features=2, cluster_std=2, random_state=2)
# split into train and test
n_train = 100
trainX, testX = X[:n_train, :], X[n_train:, :]
trainy, testy = y[:n_train], y[n_train:]
print(trainX.shape, testX.shape)
# load models in order
members = load_all_models(10)
print('Loaded %d models' % len(members))
# reverse loaded models so we build the ensemble with the last models first
members = list(reversed(members))
# evaluate different numbers of ensembles on hold out set
single_scores, ensemble_scores = list(), list()
for i in range(1, len(members)+1):
# evaluate model with i members
ensemble_score = evaluate_n_members(members, i, testX, testy)
# evaluate the i'th model standalone
testy_enc = to_categorical(testy)
_, single_score = members[i-1].evaluate(testX, testy_enc, verbose=0)
# summarize this step
print('> %d: single=%.3f, ensemble=%.3f' % (i, single_score, ensemble_score))
ensemble_scores.append(ensemble_score)
single_scores.append(single_score)
# summarize average accuracy of a single final model
print('Accuracy %.3f (%.3f)' % (mean(single_scores), std(single_scores)))
# plot score vs number of ensemble members
x_axis = [i for i in range(1, len(members)+1)]
pyplot.plot(x_axis, single_scores, marker='o', linestyle='None')
pyplot.plot(x_axis, ensemble_scores, marker='o')
pyplot.show()
Running the example first loads all 10 models into memory.
>loaded snapshot_model_1.h5 >loaded snapshot_model_2.h5 >loaded snapshot_model_3.h5 >loaded snapshot_model_4.h5 >loaded snapshot_model_5.h5 >loaded snapshot_model_6.h5 >loaded snapshot_model_7.h5 >loaded snapshot_model_8.h5 >loaded snapshot_model_9.h5 >loaded snapshot_model_10.h5 Loaded 10 models
Next, each snapshot model is evaluated on the test dataset and the accuracy is reported. This is contrasted with the accuracy of a snapshot ensemble that includes all snapshot models working backward from the end of the run including the single model.
The results show that as we work backward from the end of the run, the performance of the snapshot models gets worse, as we might expect.
Combining snapshot models into an ensemble shows that performance increases up to and including the last 3-to-5 models, reaching about 82%. This can be compared to the average performance of a snapshot model of about 80% test set accuracy.
Note, your specific results may vary given the stochastic nature of the learning algorithm. Try re-running the example a few times.
> 1: single=0.813, ensemble=0.813 > 2: single=0.814, ensemble=0.813 > 3: single=0.822, ensemble=0.822 > 4: single=0.810, ensemble=0.820 > 5: single=0.813, ensemble=0.818 > 6: single=0.811, ensemble=0.815 > 7: single=0.807, ensemble=0.813 > 8: single=0.805, ensemble=0.813 > 9: single=0.805, ensemble=0.813 > 10: single=0.790, ensemble=0.813 Accuracy 0.809 (0.008)
Finally, a line plot is created plotting the same test set accuracy scores.
We can set the performance of each individual snapshot model as a blue dot and the snapshot ensemble of increasing size (number of members) from 1 to 10 members as an orange line.
At least on this run, we can see that the snapshot ensemble quickly out-performs the final model at 82.2% with 3 members and all other saved models before performance degrades back down to about the same as the final model at 81.3%.
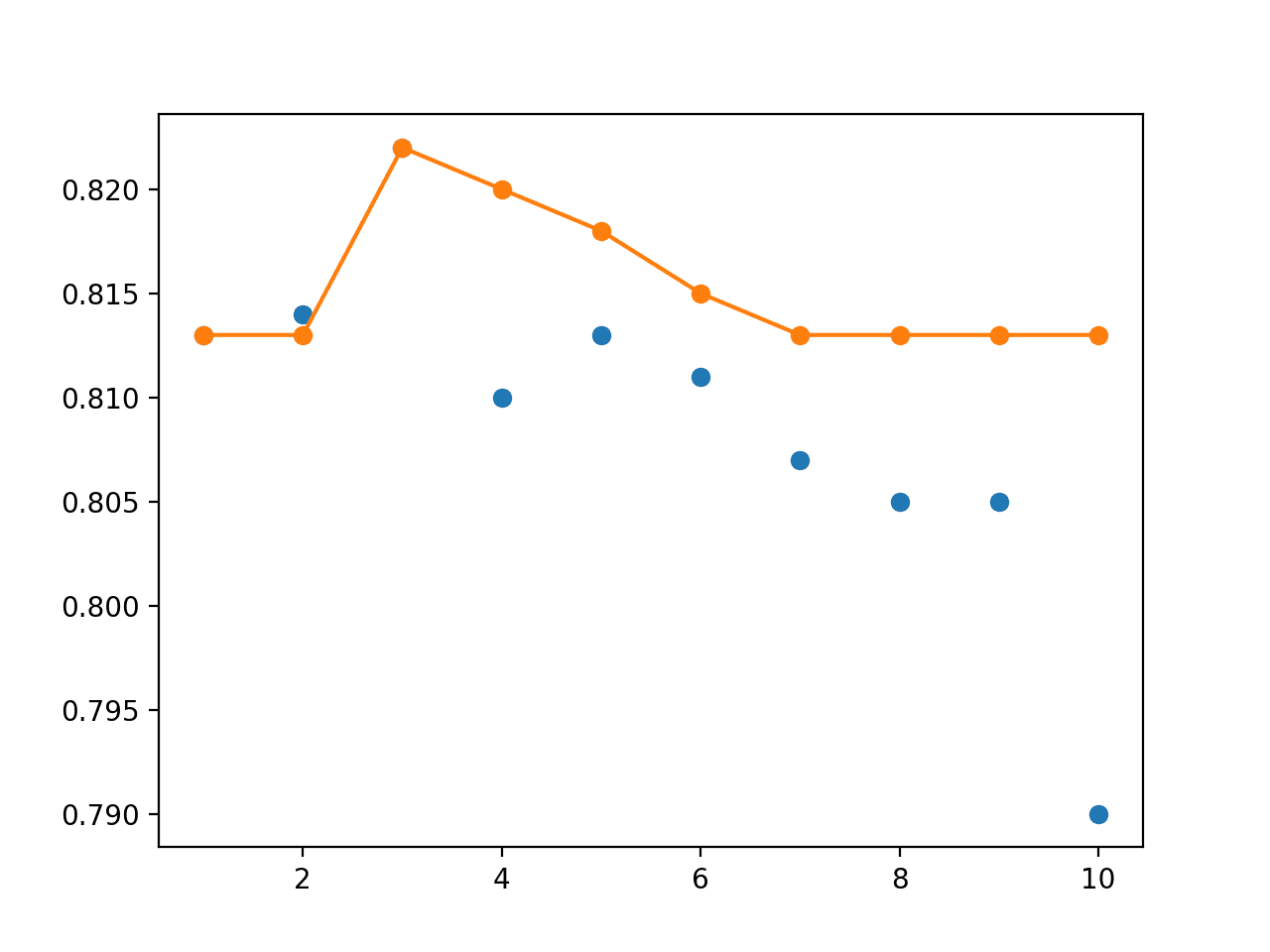
Line Plot of Single Snapshot Models (blue dots) vs Snapshot Ensembles of Varied Sized (orange line)
Extensions
This section lists some ideas for extending the tutorial that you may wish to explore.
- Vary Cycle Length. Update the example to use a shorter or longer cycle length and compare results.
- Vary Maximum Learning Rate. Update the example to use a larger or smaller maximum learning rate and compare results.
- Update Learning Rate Per Batch. Update the example to calculate the learning rate per-batch instead of per-epoch.
- Repeated Evaluation. Update the example to repeat the evaluation of the model to confirm that the approach indeed leads to an improved performance over the final model on the blobs problem.
- Cyclic Learning Rate. Update the example to use a cyclic learning rate schedule and compare results.
If you explore any of these extensions, I’d love to know.
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Papers
- Snapshot Ensembles: Train 1, get M for free, 2017.
- SGDR: Stochastic Gradient Descent with Warm Restarts, 2017.
- Cyclical Learning Rates for Training Neural Networks, 2017.
API
- Getting started with the Keras Sequential model
- Keras Core Layers API
- Keras Stochastic Gradient Descent API
- Keras Callback API
- numpy.argmax API
- sklearn.datasets.make_blobs API
Articles
- Optimization for Deep Learning Highlights in 2017.
- Cyclical Learning Rate (CLR) in Keras, GitHub.
- Snapshot Ensemble in Keras, GitHub.
- Improving the way we work with learning rate, 2017.
- The Cyclical Learning Rate technique, 2017.
Summary
In this tutorial, you discovered how to develop snapshot ensembles of models saved using an aggressive learning rate schedule over a single training run.
Specifically, you learned:
- Snapshot ensembles combine the predictions from multiple models saved during a single training run.
- Diversity in model snapshots can be achieved through the use of aggressively cycling the learning rate used during a single training run.
- How to save model snapshots during a single run and load snapshot models to make ensemble predictions.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.
The post How to Develop a Snapshot Ensemble Deep Learning Neural Network in Python With Keras appeared first on Machine Learning Mastery.
