Author: Jason Brownlee
Often, the input features for a predictive modeling task interact in unexpected and often nonlinear ways.
These interactions can be identified and modeled by a learning algorithm. Another approach is to engineer new features that expose these interactions and see if they improve model performance. Additionally, transforms like raising input variables to a power can help to better expose the important relationships between input variables and the target variable.
These features are called interaction and polynomial features and allow the use of simpler modeling algorithms as some of the complexity of interpreting the input variables and their relationships is pushed back to the data preparation stage. Sometimes these features can result in improved modeling performance, although at the cost of adding thousands or even millions of additional input variables.
In this tutorial, you will discover how to use polynomial feature transforms for feature engineering with numerical input variables.
After completing this tutorial, you will know:
- Some machine learning algorithms prefer or perform better with polynomial input features.
- How to use the polynomial features transform to create new versions of input variables for predictive modeling.
- How the degree of the polynomial impacts the number of input features created by the transform.
Let’s get started.

How to Use Polynomial Feature Transforms for Machine Learning
Photo by D Coetzee, some rights reserved.
Tutorial Overview
This tutorial is divided into five parts; they are:
- Polynomial Features
- Polynomial Feature Transform
- Sonar Dataset
- Polynomial Feature Transform Example
- Effect of Polynomial Degree
Polynomial Features
Polynomial features are those features created by raising existing features to an exponent.
For example, if a dataset had one input feature X, then a polynomial feature would be the addition of a new feature (column) where values were calculated by squaring the values in X, e.g. X^2. This process can be repeated for each input variable in the dataset, creating a transformed version of each.
As such, polynomial features are a type of feature engineering, e.g. the creation of new input features based on the existing features.
The “degree” of the polynomial is used to control the number of features added, e.g. a degree of 3 will add two new variables for each input variable. Typically a small degree is used such as 2 or 3.
Generally speaking, it is unusual to use d greater than 3 or 4 because for large values of d, the polynomial curve can become overly flexible and can take on some very strange shapes.
— Page 266, An Introduction to Statistical Learning with Applications in R, 2014.
It is also common to add new variables that represent the interaction between features, e.g a new column that represents one variable multiplied by another. This too can be repeated for each input variable creating a new “interaction” variable for each pair of input variables.
A squared or cubed version of an input variable will change the probability distribution, separating the small and large values, a separation that is increased with the size of the exponent.
This separation can help some machine learning algorithms make better predictions and is common for regression predictive modeling tasks and generally tasks that have numerical input variables.
Typically linear algorithms, such as linear regression and logistic regression, respond well to the use of polynomial input variables.
Linear regression is linear in the model parameters and adding polynomial terms to the model can be an effective way of allowing the model to identify nonlinear patterns.
— Page 11, Feature Engineering and Selection, 2019.
For example, when used as input to a linear regression algorithm, the method is more broadly referred to as polynomial regression.
Polynomial regression extends the linear model by adding extra predictors, obtained by raising each of the original predictors to a power. For example, a cubic regression uses three variables, X, X2, and X3, as predictors. This approach provides a simple way to provide a non-linear fit to data.
— Page 265, An Introduction to Statistical Learning with Applications in R, 2014.
Polynomial Feature Transform
The polynomial features transform is available in the scikit-learn Python machine learning library via the PolynomialFeatures class.
The features created include:
- The bias (the value of 1.0)
- Values raised to a power for each degree (e.g. x^1, x^2, x^3, …)
- Interactions between all pairs of features (e.g. x1 * x2, x1 * x3, …)
For example, with two input variables with values 2 and 3 and a degree of 2, the features created would be:
- 1 (the bias)
- 2^1 = 2
- 3^1 = 3
- 2^2 = 4
- 3^2 = 9
- 2 * 3 = 6
We can demonstrate this with an example:
# demonstrate the types of features created from numpy import asarray from sklearn.preprocessing import PolynomialFeatures # define the dataset data = asarray([[2,3],[2,3],[2,3]]) print(data) # perform a polynomial features transform of the dataset trans = PolynomialFeatures(degree=2) data = trans.fit_transform(data) print(data)
Running the example first reports the raw data with two features (columns) and each feature has the same value, either 2 or 3.
Then the polynomial features are created, resulting in six features, matching what was described above.
[[2 3] [2 3] [2 3]] [[1. 2. 3. 4. 6. 9.] [1. 2. 3. 4. 6. 9.] [1. 2. 3. 4. 6. 9.]]
The “degree” argument controls the number of features created and defaults to 2.
The “interaction_only” argument means that only the raw values (degree 1) and the interaction (pairs of values multiplied with each other) are included, defaulting to False.
The “include_bias” argument defaults to True to include the bias feature.
We will take a closer look at how to use the polynomial feature transforms on a real dataset.
First, let’s introduce a real dataset.
Sonar Dataset
The sonar dataset is a standard machine learning dataset for binary classification.
It involves 60 real-valued inputs and a two-class target variable. There are 208 examples in the dataset and the classes are reasonably balanced.
A baseline classification algorithm can achieve a classification accuracy of about 53.4 percent using repeated stratified 10-fold cross-validation. Top performance on this dataset is about 88 percent using repeated stratified 10-fold cross-validation.
The dataset describes radar returns of rocks or simulated mines.
You can learn more about the dataset from here:
No need to download the dataset; we will download it automatically from our worked examples.
First, let’s load and summarize the dataset. The complete example is listed below.
# load and summarize the sonar dataset from pandas import read_csv from pandas.plotting import scatter_matrix from matplotlib import pyplot # Load dataset url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv" dataset = read_csv(url, header=None) # summarize the shape of the dataset print(dataset.shape) # summarize each variable print(dataset.describe()) # histograms of the variables dataset.hist() pyplot.show()
Running the example first summarizes the shape of the loaded dataset.
This confirms the 60 input variables, one output variable, and 208 rows of data.
A statistical summary of the input variables is provided showing that values are numeric and range approximately from 0 to 1.
(208, 61)
0 1 2 ... 57 58 59
count 208.000000 208.000000 208.000000 ... 208.000000 208.000000 208.000000
mean 0.029164 0.038437 0.043832 ... 0.007949 0.007941 0.006507
std 0.022991 0.032960 0.038428 ... 0.006470 0.006181 0.005031
min 0.001500 0.000600 0.001500 ... 0.000300 0.000100 0.000600
25% 0.013350 0.016450 0.018950 ... 0.003600 0.003675 0.003100
50% 0.022800 0.030800 0.034300 ... 0.005800 0.006400 0.005300
75% 0.035550 0.047950 0.057950 ... 0.010350 0.010325 0.008525
max 0.137100 0.233900 0.305900 ... 0.044000 0.036400 0.043900
[8 rows x 60 columns]
Finally, a histogram is created for each input variable.
If we ignore the clutter of the plots and focus on the histograms themselves, we can see that many variables have a skewed distribution.
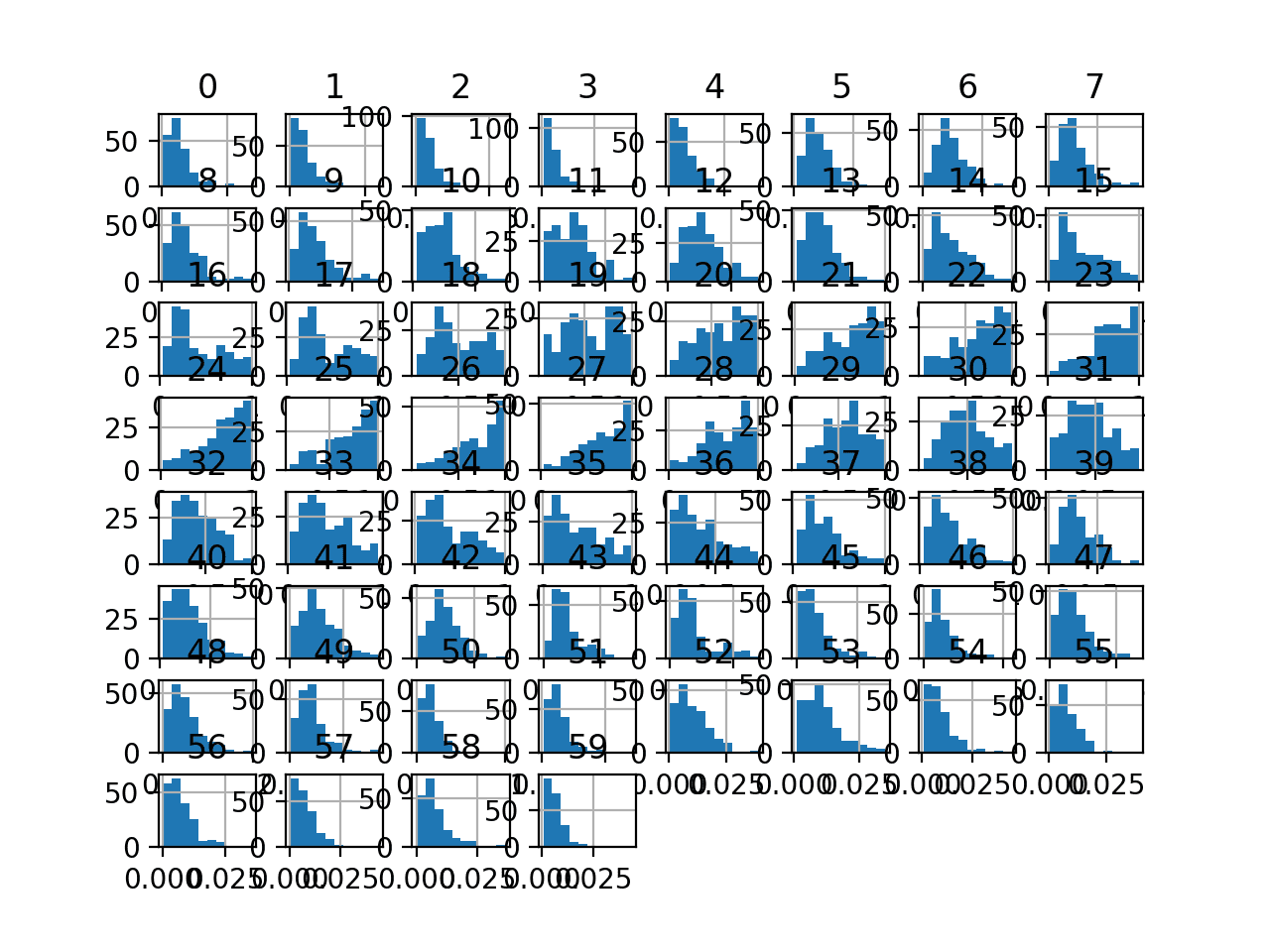
Histogram Plots of Input Variables for the Sonar Binary Classification Dataset
Next, let’s fit and evaluate a machine learning model on the raw dataset.
We will use a k-nearest neighbor algorithm with default hyperparameters and evaluate it using repeated stratified k-fold cross-validation. The complete example is listed below.
# evaluate knn on the raw sonar dataset
from numpy import mean
from numpy import std
from pandas import read_csv
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import LabelEncoder
from matplotlib import pyplot
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv"
dataset = read_csv(url, header=None)
data = dataset.values
# separate into input and output columns
X, y = data[:, :-1], data[:, -1]
# ensure inputs are floats and output is an integer label
X = X.astype('float32')
y = LabelEncoder().fit_transform(y.astype('str'))
# define and configure the model
model = KNeighborsClassifier()
# evaluate the model
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
# report model performance
print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores)))
Running the example evaluates a KNN model on the raw sonar dataset.
We can see that the model achieved a mean classification accuracy of about 79.7 percent, showing that it has skill (better than 53.4 percent) and is in the ball-park of good performance (88 percent).
Accuracy: 0.797 (0.073)
Next, let’s explore a polynomial features transform of the dataset.
Polynomial Feature Transform Example
We can apply the polynomial features transform to the Sonar dataset directly.
In this case, we will use a degree of 3.
... # perform a polynomial features transform of the dataset trans = PolynomialFeatures(degree=3) data = trans.fit_transform(data)
Let’s try it on our sonar dataset.
The complete example of creating a polynomial features transform of the sonar dataset and summarizing the created features is below.
# visualize a polynomial features transform of the sonar dataset from pandas import read_csv from pandas import DataFrame from pandas.plotting import scatter_matrix from sklearn.preprocessing import PolynomialFeatures from matplotlib import pyplot # load dataset url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv" dataset = read_csv(url, header=None) # retrieve just the numeric input values data = dataset.values[:, :-1] # perform a polynomial features transform of the dataset trans = PolynomialFeatures(degree=3) data = trans.fit_transform(data) # convert the array back to a dataframe dataset = DataFrame(data) # summarize print(dataset.shape)
Running the example performs the polynomial features transform on the sonar dataset.
We can see that our features increased from 61 (60 input features) for the raw dataset to 39,711 features (39,710 input features).
(208, 39711)
Next, let’s evaluate the same KNN model as the previous section, but in this case on a polynomial features transform of the dataset.
The complete example is listed below.
# evaluate knn on the sonar dataset with polynomial features transform
from numpy import mean
from numpy import std
from pandas import read_csv
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import Pipeline
from matplotlib import pyplot
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv"
dataset = read_csv(url, header=None)
data = dataset.values
# separate into input and output columns
X, y = data[:, :-1], data[:, -1]
# ensure inputs are floats and output is an integer label
X = X.astype('float32')
y = LabelEncoder().fit_transform(y.astype('str'))
# define the pipeline
trans = PolynomialFeatures(degree=3)
model = KNeighborsClassifier()
pipeline = Pipeline(steps=[('t', trans), ('m', model)])
# evaluate the pipeline
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(pipeline, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
# report pipeline performance
print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores)))
Running the example, we can see that the polynomial features transform results in a lift in performance from 79.7 percent accuracy without the transform to about 80.0 percent with the transform.
Accuracy: 0.800 (0.077)
Next, let’s explore the effect of different scaling ranges.
Effect of Polynomial Degree
The degree of the polynomial dramatically increases the number of input features.
To get an idea of how much this impacts the number of features, we can perform the transform with a range of different degrees and compare the number of features in the dataset.
The complete example is listed below.
# compare the effect of the degree on the number of created features
from pandas import read_csv
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import PolynomialFeatures
from matplotlib import pyplot
# get the dataset
def get_dataset():
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv"
dataset = read_csv(url, header=None)
data = dataset.values
# separate into input and output columns
X, y = data[:, :-1], data[:, -1]
# ensure inputs are floats and output is an integer label
X = X.astype('float32')
y = LabelEncoder().fit_transform(y.astype('str'))
return X, y
# define dataset
X, y = get_dataset()
# calculate change in number of features
num_features = list()
degress = [i for i in range(1, 6)]
for d in degress:
# create transform
trans = PolynomialFeatures(degree=d)
# fit and transform
data = trans.fit_transform(X)
# record number of features
num_features.append(data.shape[1])
# summarize
print('Degree: %d, Features: %d' % (d, data.shape[1]))
# plot degree vs number of features
pyplot.plot(degress, num_features)
pyplot.show()
Running the example first reports the degree from 1 to 5 and the number of features in the dataset.
We can see that a degree of 1 has no effect and that the number of features dramatically increases from 2 through to 5.
This highlights that for anything other than very small datasets, a degree of 2 or 3 should be used to avoid a dramatic increase in input variables.
Degree: 1, Features: 61 Degree: 2, Features: 1891 Degree: 3, Features: 39711 Degree: 4, Features: 635376 Degree: 5, Features: 8259888
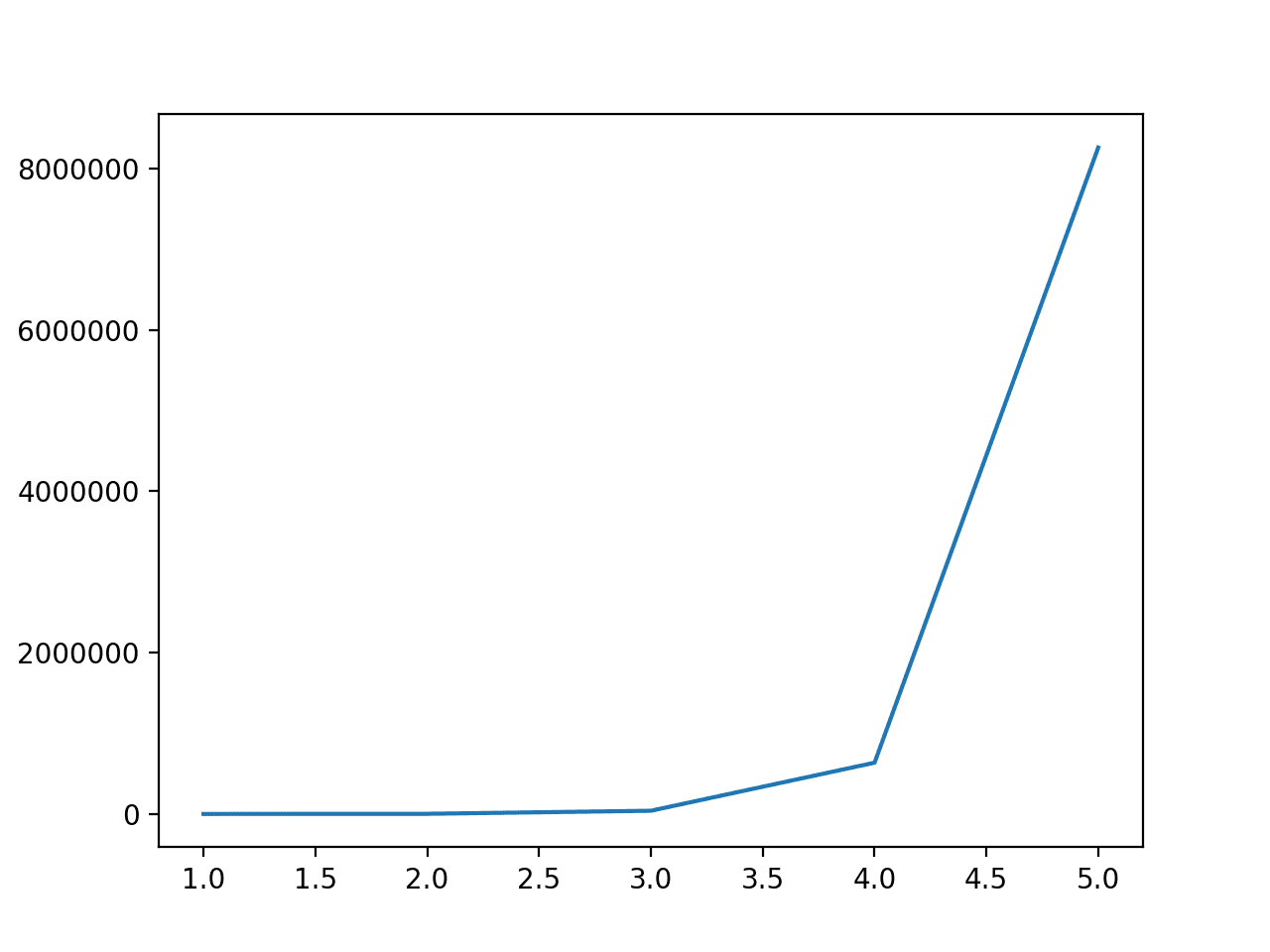
Line Plot of the Degree vs. the Number of Input Features for the Polynomial Feature Transform
More features may result in more overfitting, and in turn, worse results.
It may be a good idea to treat the degree for the polynomial features transform as a hyperparameter and test different values for your dataset.
The example below explores degree values from 1 to 4 and evaluates their effect on classification accuracy with the chosen model.
# explore the effect of degree on accuracy for the polynomial features transform
from numpy import mean
from numpy import std
from pandas import read_csv
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import PolynomialFeatures
from sklearn.preprocessing import LabelEncoder
from sklearn.pipeline import Pipeline
from matplotlib import pyplot
# get the dataset
def get_dataset():
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv"
dataset = read_csv(url, header=None)
data = dataset.values
# separate into input and output columns
X, y = data[:, :-1], data[:, -1]
# ensure inputs are floats and output is an integer label
X = X.astype('float32')
y = LabelEncoder().fit_transform(y.astype('str'))
return X, y
# get a list of models to evaluate
def get_models():
models = dict()
for d in range(1,5):
# define the pipeline
trans = PolynomialFeatures(degree=d)
model = KNeighborsClassifier()
models[str(d)] = Pipeline(steps=[('t', trans), ('m', model)])
return models
# evaluate a give model using cross-validation
def evaluate_model(model):
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
return scores
# define dataset
X, y = get_dataset()
# get the models to evaluate
models = get_models()
# evaluate the models and store results
results, names = list(), list()
for name, model in models.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print('>%s %.3f (%.3f)' % (name, mean(scores), std(scores)))
# plot model performance for comparison
pyplot.boxplot(results, labels=names, showmeans=True)
pyplot.show()
Running the example reports the mean classification accuracy for each polynomial degree.
In this case, we can see that performance is generally worse than no transform (degree 1) except for a degree 3.
It might be interesting to explore scaling the data before or after performing the transform to see how it impacts model performance.
>1 0.797 (0.073) >2 0.793 (0.085) >3 0.800 (0.077) >4 0.795 (0.079)
Box and whisker plots are created to summarize the classification accuracy scores for each polynomial degree.
We can see that performance remains flat, perhaps with the first signs of overfitting with a degree of 4.
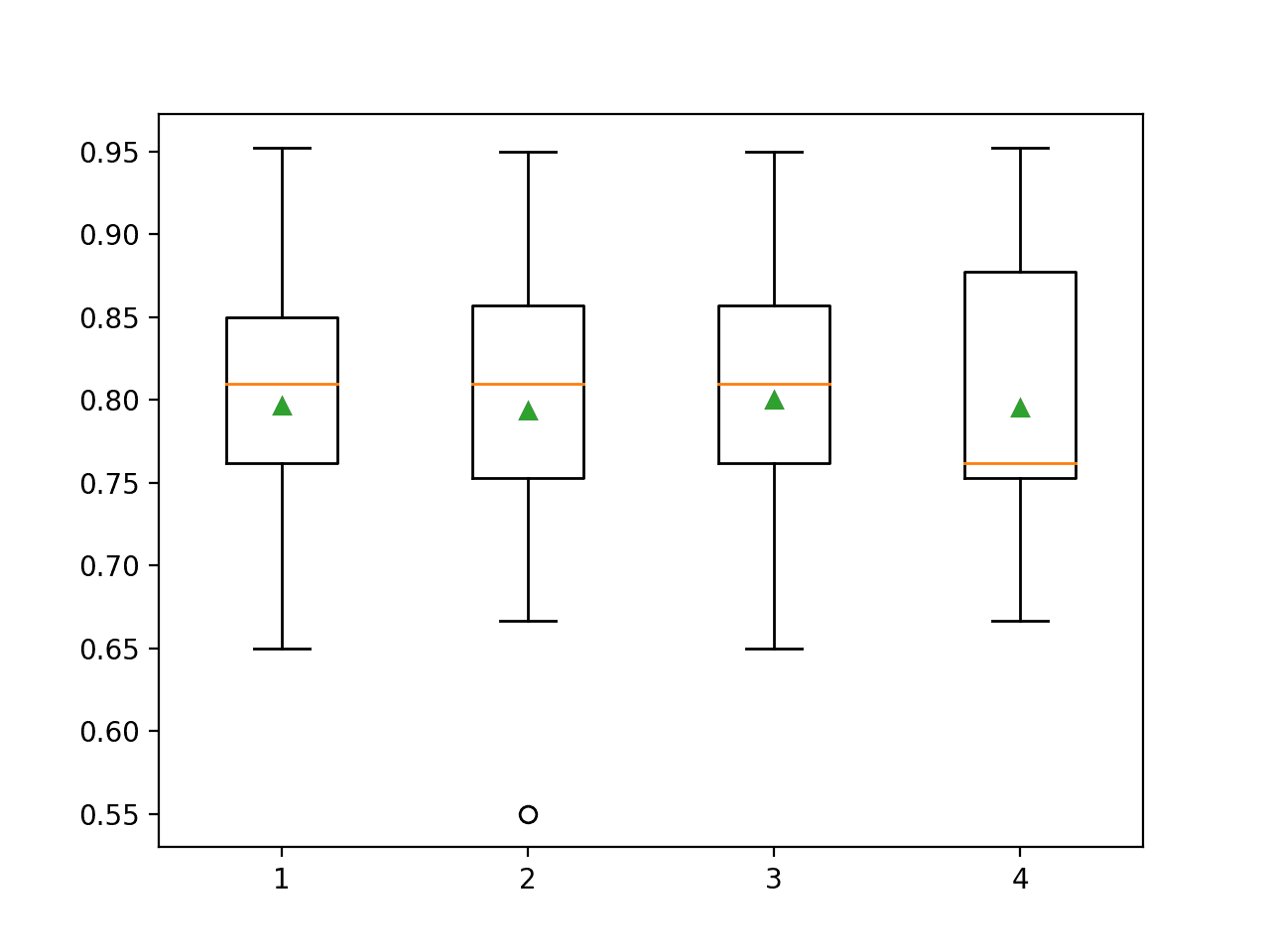
Box Plots of Degree for the Polynomial Feature Transform vs. Classification Accuracy of KNN on the Sonar Dataset
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Books
- An Introduction to Statistical Learning with Applications in R, 2014.
- Feature Engineering and Selection, 2019.
APIs
Articles
Summary
In this tutorial, you discovered how to use polynomial feature transforms for feature engineering with numerical input variables.
Specifically, you learned:
- Some machine learning algorithms prefer or perform better with polynomial input features.
- How to use the polynomial features transform to create new versions of input variables for predictive modeling.
- How the degree of the polynomial impacts the number of input features created by the transform.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.
The post How to Use Polynomial Feature Transforms for Machine Learning appeared first on Machine Learning Mastery.
