Author: Jason Brownlee
Gradient descent is an optimization algorithm that follows the negative gradient of an objective function in order to locate the minimum of the function.
A limitation of gradient descent is that it uses the same step size (learning rate) for each input variable. AdaGradn and RMSProp are extensions to gradient descent that add a self-adaptive learning rate for each parameter for the objective function.
Adadelta can be considered a further extension of gradient descent that builds upon AdaGrad and RMSProp and changes the calculation of the custom step size so that the units are consistent and in turn no longer requires an initial learning rate hyperparameter.
In this tutorial, you will discover how to develop the gradient descent with Adadelta optimization algorithm from scratch.
After completing this tutorial, you will know:
- Gradient descent is an optimization algorithm that uses the gradient of the objective function to navigate the search space.
- Gradient descent can be updated to use an automatically adaptive step size for each input variable using a decaying average of partial derivatives, called Adadelta.
- How to implement the Adadelta optimization algorithm from scratch and apply it to an objective function and evaluate the results.
Let’s get started.

Gradient Descent With Adadelta from Scratch
Photo by Robert Minkler, some rights reserved.
Tutorial Overview
This tutorial is divided into three parts; they are:
- Gradient Descent
- Adadelta Algorithm
- Gradient Descent With Adadelta
- Two-Dimensional Test Problem
- Gradient Descent Optimization With Adadelta
- Visualization of Adadelta
Gradient Descent
Gradient descent is an optimization algorithm.
It is technically referred to as a first-order optimization algorithm as it explicitly makes use of the first-order derivative of the target objective function.
First-order methods rely on gradient information to help direct the search for a minimum …
— Page 69, Algorithms for Optimization, 2019.
The first order derivative, or simply the “derivative,” is the rate of change or slope of the target function at a specific point, e.g. for a specific input.
If the target function takes multiple input variables, it is referred to as a multivariate function and the input variables can be thought of as a vector. In turn, the derivative of a multivariate target function may also be taken as a vector and is referred to generally as the gradient.
- Gradient: First-order derivative for a multivariate objective function.
The derivative or the gradient points in the direction of the steepest ascent of the target function for a specific input.
Gradient descent refers to a minimization optimization algorithm that follows the negative of the gradient downhill of the target function to locate the minimum of the function.
The gradient descent algorithm requires a target function that is being optimized and the derivative function for the objective function. The target function f() returns a score for a given set of inputs, and the derivative function f'() gives the derivative of the target function for a given set of inputs.
The gradient descent algorithm requires a starting point (x) in the problem, such as a randomly selected point in the input space.
The derivative is then calculated and a step is taken in the input space that is expected to result in a downhill movement in the target function, assuming we are minimizing the target function.
A downhill movement is made by first calculating how far to move in the input space, calculated as the steps size (called alpha or the learning rate) multiplied by the gradient. This is then subtracted from the current point, ensuring we move against the gradient, or down the target function.
- x = x – step_size * f'(x)
The steeper the objective function at a given point, the larger the magnitude of the gradient, and in turn, the larger the step taken in the search space. The size of the step taken is scaled using a step size hyperparameter.
- Step Size (alpha): Hyperparameter that controls how far to move in the search space against the gradient each iteration of the algorithm.
If the step size is too small, the movement in the search space will be small and the search will take a long time. If the step size is too large, the search may bounce around the search space and skip over the optima.
Now that we are familiar with the gradient descent optimization algorithm, let’s take a look at Adadelta.
Adadelta Algorithm
Adadelta (or “ADADELTA”) is an extension to the gradient descent optimization algorithm.
The algorithm was described in the 2012 paper by Matthew Zeiler titled “ADADELTA: An Adaptive Learning Rate Method.”
Adadelta is designed to accelerate the optimization process, e.g. decrease the number of function evaluations required to reach the optima, or to improve the capability of the optimization algorithm, e.g. result in a better final result.
It is best understood as an extension of the AdaGrad and RMSProp algorithms.
AdaGrad is an extension of gradient descent that calculates a step size (learning rate) for each parameter for the objective function each time an update is made. The step size is calculated by first summing the partial derivatives for the parameter seen so far during the search, then dividing the initial step size hyperparameter by the square root of the sum of the squared partial derivatives.
The calculation of the custom step size for one parameter with AdaGrad is as follows:
- cust_step_size(t+1) = step_size / (1e-8 + sqrt(s(t)))
Where cust_step_size(t+1) is the calculated step size for an input variable for a given point during the search, step_size is the initial step size, sqrt() is the square root operation, and s(t) is the sum of the squared partial derivatives for the input variable seen during the search so far (including the current iteration).
RMSProp can be thought of as an extension of AdaGrad in that it uses a decaying average or moving average of the partial derivatives instead of the sum in the calculation of the step size for each parameter. This is achieved by adding a new hyperparameter “rho” that acts like a momentum for the partial derivatives.
The calculation of the decaying moving average squared partial derivative for one parameter is as follows:
- s(t+1) = (s(t) * rho) + (f'(x(t))^2 * (1.0-rho))
Where s(t+1) is the mean squared partial derivative for one parameter for the current iteration of the algorithm, s(t) is the decaying moving average squared partial derivative for the previous iteration, f'(x(t))^2 is the squared partial derivative for the current parameter, and rho is a hyperparameter, typically with the value of 0.9 like momentum.
Adadelta is a further extension of RMSProp designed to improve the convergence of the algorithm and to remove the need for a manually specified initial learning rate.
The idea presented in this paper was derived from ADAGRAD in order to improve upon the two main drawbacks of the method: 1) the continual decay of learning rates throughout training, and 2) the need for a manually selected global learning rate.
— ADADELTA: An Adaptive Learning Rate Method, 2012.
The decaying moving average of the squared partial derivative is calculated for each parameter, as with RMSProp. The key difference is in the calculation of the step size for a parameter that uses the decaying average of the delta or change in parameter.
This choice of numerator was to ensure that both parts of the calculation have the same units.
After independently deriving the RMSProp update, the authors noticed that the units in the update equations for gradient descent, momentum and Adagrad do not match. To fix this, they use an exponentially decaying average of the square updates
— Pages 78-79, Algorithms for Optimization, 2019.
First, the custom step size is calculated as the square root of the decaying moving average of the change in the delta divided by the square root of the decaying moving average of the squared partial derivatives.
- cust_step_size(t+1) = (ep + sqrt(delta(t))) / (ep + sqrt(s(t)))
Where cust_step_size(t+1) is the custom step size for a parameter for a given update, ep is a hyperparameter that is added to the numerator and denominator to avoid a divide by zero error, delta(t) is the decaying moving average of the squared change to the parameter (calculated in the last iteration), and s(t) is the decaying moving average of the squared partial derivative (calculated in the current iteration).
The ep hyperparameter is set to a small value such as 1e-3 or 1e-8. In addition to avoiding a divide by zero error, it also helps with the first step of the algorithm when the decaying moving average squared change and decaying moving average squared gradient are zero.
Next, the change to the parameter is calculated as the custom step size multiplied by the partial derivative
- change(t+1) = cust_step_size(t+1) * f'(x(t))
Next, the decaying average of the squared change to the parameter is updated.
- delta(t+1) = (delta(t) * rho) + (change(t+1)^2 * (1.0-rho))
Where delta(t+1) is the decaying average of the change to the variable to be used in the next iteration, change(t+1) was calculated in the step before and rho is a hyperparameter that acts like momentum and has a value like 0.9.
Finally, the new value for the variable is calculated using the change.
- x(t+1) = x(t) – change(t+1)
This process is then repeated for each variable for the objective function, then the entire process is repeated to navigate the search space for a fixed number of algorithm iterations.
Now that we are familiar with the Adadelta algorithm, let’s explore how we might implement it and evaluate its performance.
Gradient Descent With Adadelta
In this section, we will explore how to implement the gradient descent optimization algorithm with Adadelta.
Two-Dimensional Test Problem
First, let’s define an optimization function.
We will use a simple two-dimensional function that squares the input of each dimension and define the range of valid inputs from -1.0 to 1.0.
The objective() function below implements this function
# objective function def objective(x, y): return x**2.0 + y**2.0
We can create a three-dimensional plot of the dataset to get a feeling for the curvature of the response surface.
The complete example of plotting the objective function is listed below.
# 3d plot of the test function from numpy import arange from numpy import meshgrid from matplotlib import pyplot # objective function def objective(x, y): return x**2.0 + y**2.0 # define range for input r_min, r_max = -1.0, 1.0 # sample input range uniformly at 0.1 increments xaxis = arange(r_min, r_max, 0.1) yaxis = arange(r_min, r_max, 0.1) # create a mesh from the axis x, y = meshgrid(xaxis, yaxis) # compute targets results = objective(x, y) # create a surface plot with the jet color scheme figure = pyplot.figure() axis = figure.gca(projection='3d') axis.plot_surface(x, y, results, cmap='jet') # show the plot pyplot.show()
Running the example creates a three dimensional surface plot of the objective function.
We can see the familiar bowl shape with the global minima at f(0, 0) = 0.
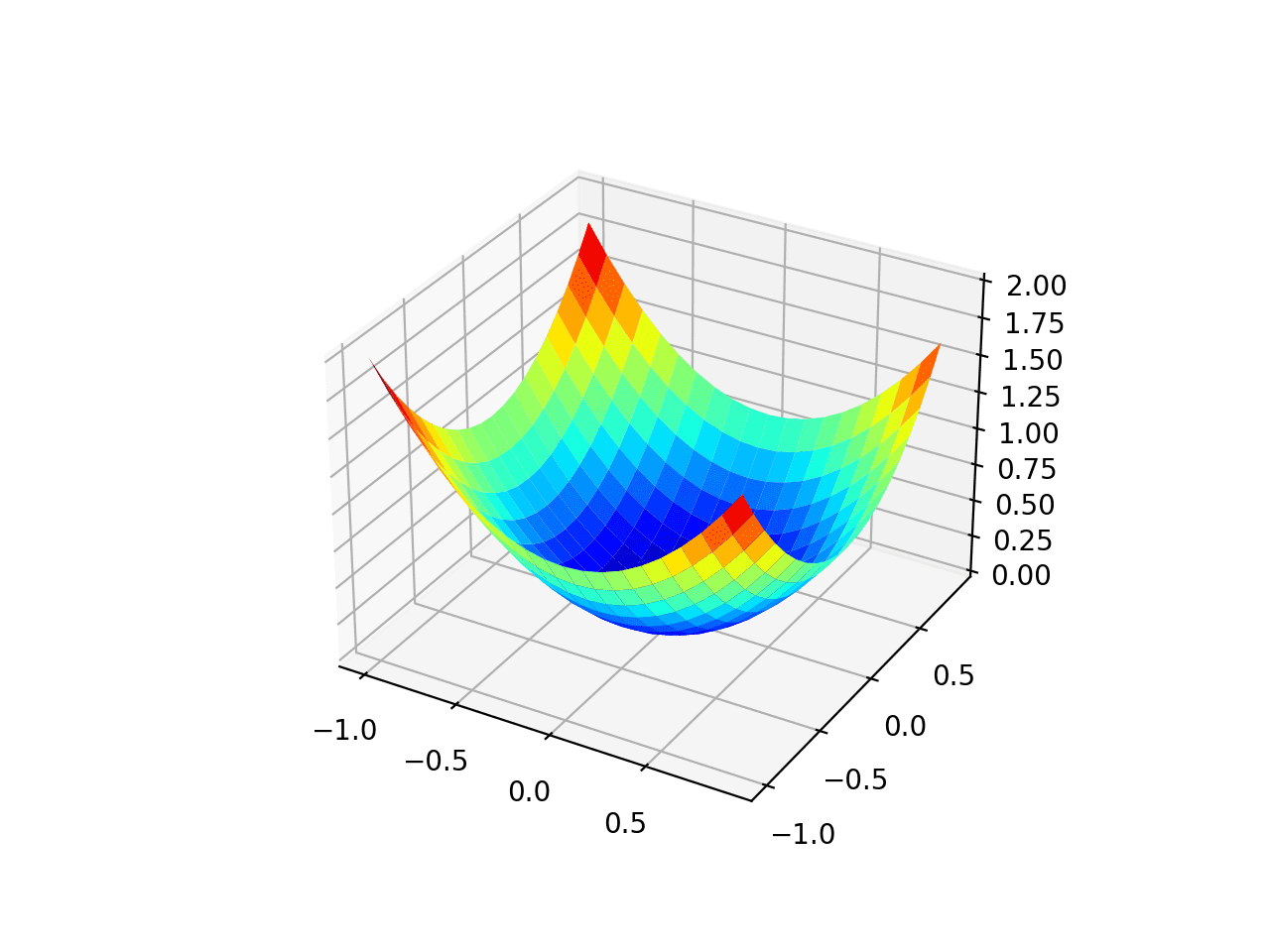
Three-Dimensional Plot of the Test Objective Function
We can also create a two-dimensional plot of the function. This will be helpful later when we want to plot the progress of the search.
The example below creates a contour plot of the objective function.
# contour plot of the test function from numpy import asarray from numpy import arange from numpy import meshgrid from matplotlib import pyplot # objective function def objective(x, y): return x**2.0 + y**2.0 # define range for input bounds = asarray([[-1.0, 1.0], [-1.0, 1.0]]) # sample input range uniformly at 0.1 increments xaxis = arange(bounds[0,0], bounds[0,1], 0.1) yaxis = arange(bounds[1,0], bounds[1,1], 0.1) # create a mesh from the axis x, y = meshgrid(xaxis, yaxis) # compute targets results = objective(x, y) # create a filled contour plot with 50 levels and jet color scheme pyplot.contourf(x, y, results, levels=50, cmap='jet') # show the plot pyplot.show()
Running the example creates a two-dimensional contour plot of the objective function.
We can see the bowl shape compressed to contours shown with a color gradient. We will use this plot to plot the specific points explored during the progress of the search.
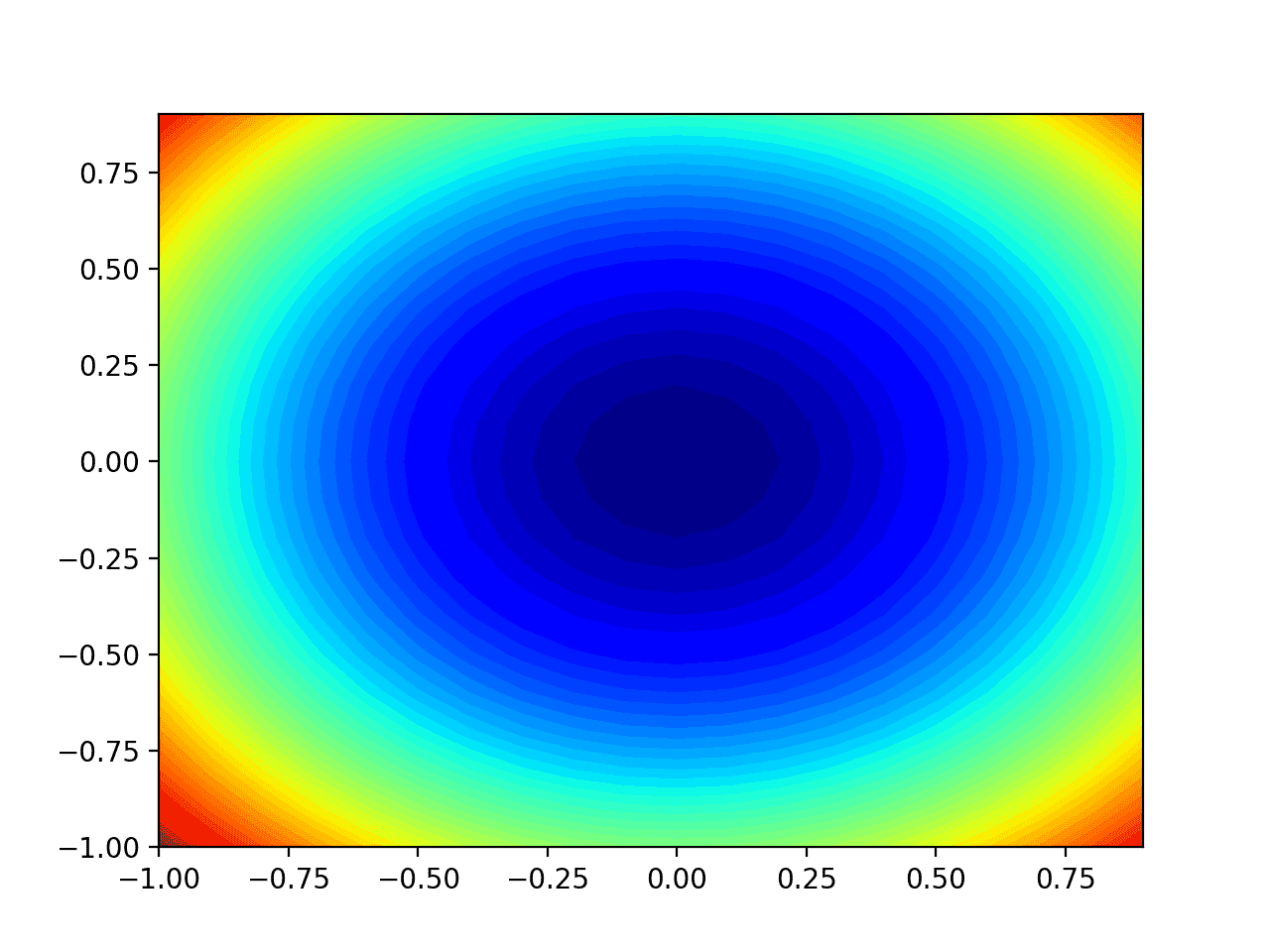
Two-Dimensional Contour Plot of the Test Objective Function
Now that we have a test objective function, let’s look at how we might implement the Adadelta optimization algorithm.
Gradient Descent Optimization With Adadelta
We can apply the gradient descent with Adadelta to the test problem.
First, we need a function that calculates the derivative for this function.
- f(x) = x^2
- f'(x) = x * 2
The derivative of x^2 is x * 2 in each dimension. The derivative() function implements this below.
# derivative of objective function def derivative(x, y): return asarray([x * 2.0, y * 2.0])
Next, we can implement gradient descent optimization.
First, we can select a random point in the bounds of the problem as a starting point for the search.
This assumes we have an array that defines the bounds of the search with one row for each dimension and the first column defines the minimum and the second column defines the maximum of the dimension.
... # generate an initial point solution = bounds[:, 0] + rand(len(bounds)) * (bounds[:, 1] - bounds[:, 0])
Next, we need to initialize the decaying average of the squared partial derivatives and squared change for each dimension to 0.0 values.
... # list of the average square gradients for each variable sq_grad_avg = [0.0 for _ in range(bounds.shape[0])] # list of the average parameter updates sq_para_avg = [0.0 for _ in range(bounds.shape[0])]
We can then enumerate a fixed number of iterations of the search optimization algorithm defined by a “n_iter” hyperparameter.
... # run the gradient descent for it in range(n_iter): ...
The first step is to calculate the gradient for the current solution using the derivative() function.
... # calculate gradient gradient = derivative(solution[0], solution[1])
We then need to calculate the square of the partial derivative and update the decaying moving average of the squared partial derivatives with the “rho” hyperparameter.
... # update the average of the squared partial derivatives for i in range(gradient.shape[0]): # calculate the squared gradient sg = gradient[i]**2.0 # update the moving average of the squared gradient sq_grad_avg[i] = (sq_grad_avg[i] * rho) + (sg * (1.0-rho))
We can then use the decaying moving average of the squared partial derivatives and gradient to calculate the step size for the next point. We will do this one variable at a time.
... # build solution new_solution = list() for i in range(solution.shape[0]): ...
First, we will calculate the custom step size for this variable on this iteration using the decaying moving average of the squared changes and squared partial derivatives, as well as the “ep” hyperparameter.
... # calculate the step size for this variable alpha = (ep + sqrt(sq_para_avg[i])) / (ep + sqrt(sq_grad_avg[i]))
Next, we can use the custom step size and partial derivative to calculate the change to the variable.
... # calculate the change change = alpha * gradient[i]
We can then use the change to update the decaying moving average of the squared change using the “rho” hyperparameter.
... # update the moving average of squared parameter changes sq_para_avg[i] = (sq_para_avg[i] * rho) + (change**2.0 * (1.0-rho))
Finally, we can change the variable and store the result before moving on to the next variable.
... # calculate the new position in this variable value = solution[i] - change # store this variable new_solution.append(value)
This new solution can then be evaluated using the objective() function and the performance of the search can be reported.
...
# evaluate candidate point
solution = asarray(new_solution)
solution_eval = objective(solution[0], solution[1])
# report progress
print('>%d f(%s) = %.5f' % (it, solution, solution_eval))
And that’s it.
We can tie all of this together into a function named adadelta() that takes the names of the objective function and the derivative function, an array with the bounds of the domain and hyperparameter values for the total number of algorithm iterations and rho, and returns the final solution and its evaluation.
The ep hyperparameter can also be taken as an argument, although has a sensible default value of 1e-3.
This complete function is listed below.
# gradient descent algorithm with adadelta
def adadelta(objective, derivative, bounds, n_iter, rho, ep=1e-3):
# generate an initial point
solution = bounds[:, 0] + rand(len(bounds)) * (bounds[:, 1] - bounds[:, 0])
# list of the average square gradients for each variable
sq_grad_avg = [0.0 for _ in range(bounds.shape[0])]
# list of the average parameter updates
sq_para_avg = [0.0 for _ in range(bounds.shape[0])]
# run the gradient descent
for it in range(n_iter):
# calculate gradient
gradient = derivative(solution[0], solution[1])
# update the average of the squared partial derivatives
for i in range(gradient.shape[0]):
# calculate the squared gradient
sg = gradient[i]**2.0
# update the moving average of the squared gradient
sq_grad_avg[i] = (sq_grad_avg[i] * rho) + (sg * (1.0-rho))
# build a solution one variable at a time
new_solution = list()
for i in range(solution.shape[0]):
# calculate the step size for this variable
alpha = (ep + sqrt(sq_para_avg[i])) / (ep + sqrt(sq_grad_avg[i]))
# calculate the change
change = alpha * gradient[i]
# update the moving average of squared parameter changes
sq_para_avg[i] = (sq_para_avg[i] * rho) + (change**2.0 * (1.0-rho))
# calculate the new position in this variable
value = solution[i] - change
# store this variable
new_solution.append(value)
# evaluate candidate point
solution = asarray(new_solution)
solution_eval = objective(solution[0], solution[1])
# report progress
print('>%d f(%s) = %.5f' % (it, solution, solution_eval))
return [solution, solution_eval]
Note: we have intentionally used lists and imperative coding style instead of vectorized operations for readability. Feel free to adapt the implementation to a vectorization implementation with NumPy arrays for better performance.
We can then define our hyperparameters and call the adadelta() function to optimize our test objective function.
In this case, we will use 120 iterations of the algorithm and a value of 0.99 for the rho hyperparameter, chosen after a little trial and error.
...
# seed the pseudo random number generator
seed(1)
# define range for input
bounds = asarray([[-1.0, 1.0], [-1.0, 1.0]])
# define the total iterations
n_iter = 120
# momentum for adadelta
rho = 0.99
# perform the gradient descent search with adadelta
best, score = adadelta(objective, derivative, bounds, n_iter, rho)
print('Done!')
print('f(%s) = %f' % (best, score))
Tying all of this together, the complete example of gradient descent optimization with Adadelta is listed below.
# gradient descent optimization with adadelta for a two-dimensional test function
from math import sqrt
from numpy import asarray
from numpy.random import rand
from numpy.random import seed
# objective function
def objective(x, y):
return x**2.0 + y**2.0
# derivative of objective function
def derivative(x, y):
return asarray([x * 2.0, y * 2.0])
# gradient descent algorithm with adadelta
def adadelta(objective, derivative, bounds, n_iter, rho, ep=1e-3):
# generate an initial point
solution = bounds[:, 0] + rand(len(bounds)) * (bounds[:, 1] - bounds[:, 0])
# list of the average square gradients for each variable
sq_grad_avg = [0.0 for _ in range(bounds.shape[0])]
# list of the average parameter updates
sq_para_avg = [0.0 for _ in range(bounds.shape[0])]
# run the gradient descent
for it in range(n_iter):
# calculate gradient
gradient = derivative(solution[0], solution[1])
# update the average of the squared partial derivatives
for i in range(gradient.shape[0]):
# calculate the squared gradient
sg = gradient[i]**2.0
# update the moving average of the squared gradient
sq_grad_avg[i] = (sq_grad_avg[i] * rho) + (sg * (1.0-rho))
# build a solution one variable at a time
new_solution = list()
for i in range(solution.shape[0]):
# calculate the step size for this variable
alpha = (ep + sqrt(sq_para_avg[i])) / (ep + sqrt(sq_grad_avg[i]))
# calculate the change
change = alpha * gradient[i]
# update the moving average of squared parameter changes
sq_para_avg[i] = (sq_para_avg[i] * rho) + (change**2.0 * (1.0-rho))
# calculate the new position in this variable
value = solution[i] - change
# store this variable
new_solution.append(value)
# evaluate candidate point
solution = asarray(new_solution)
solution_eval = objective(solution[0], solution[1])
# report progress
print('>%d f(%s) = %.5f' % (it, solution, solution_eval))
return [solution, solution_eval]
# seed the pseudo random number generator
seed(1)
# define range for input
bounds = asarray([[-1.0, 1.0], [-1.0, 1.0]])
# define the total iterations
n_iter = 120
# momentum for adadelta
rho = 0.99
# perform the gradient descent search with adadelta
best, score = adadelta(objective, derivative, bounds, n_iter, rho)
print('Done!')
print('f(%s) = %f' % (best, score))
Running the example applies the Adadelta optimization algorithm to our test problem and reports performance of the search for each iteration of the algorithm.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that a near optimal solution was found after perhaps 105 iterations of the search, with input values near 0.0 and 0.0, evaluating to 0.0.
... >100 f([-1.45142626e-07 2.71163181e-03]) = 0.00001 >101 f([-1.24898699e-07 2.56875692e-03]) = 0.00001 >102 f([-1.07454197e-07 2.43328237e-03]) = 0.00001 >103 f([-9.24253035e-08 2.30483111e-03]) = 0.00001 >104 f([-7.94803792e-08 2.18304501e-03]) = 0.00000 >105 f([-6.83329263e-08 2.06758392e-03]) = 0.00000 >106 f([-5.87354975e-08 1.95812477e-03]) = 0.00000 >107 f([-5.04744185e-08 1.85436071e-03]) = 0.00000 >108 f([-4.33652179e-08 1.75600036e-03]) = 0.00000 >109 f([-3.72486699e-08 1.66276699e-03]) = 0.00000 >110 f([-3.19873691e-08 1.57439783e-03]) = 0.00000 >111 f([-2.74627662e-08 1.49064334e-03]) = 0.00000 >112 f([-2.3572602e-08 1.4112666e-03]) = 0.00000 >113 f([-2.02286891e-08 1.33604264e-03]) = 0.00000 >114 f([-1.73549914e-08 1.26475787e-03]) = 0.00000 >115 f([-1.48859650e-08 1.19720951e-03]) = 0.00000 >116 f([-1.27651224e-08 1.13320504e-03]) = 0.00000 >117 f([-1.09437923e-08 1.07256172e-03]) = 0.00000 >118 f([-9.38004754e-09 1.01510604e-03]) = 0.00000 >119 f([-8.03777865e-09 9.60673346e-04]) = 0.00000 Done! f([-8.03777865e-09 9.60673346e-04]) = 0.000001
Visualization of Adadelta
We can plot the progress of the Adadelta search on a contour plot of the domain.
This can provide an intuition for the progress of the search over the iterations of the algorithm.
We must update the adadelta() function to maintain a list of all solutions found during the search, then return this list at the end of the search.
The updated version of the function with these changes is listed below.
# gradient descent algorithm with adadelta
def adadelta(objective, derivative, bounds, n_iter, rho, ep=1e-3):
# track all solutions
solutions = list()
# generate an initial point
solution = bounds[:, 0] + rand(len(bounds)) * (bounds[:, 1] - bounds[:, 0])
# list of the average square gradients for each variable
sq_grad_avg = [0.0 for _ in range(bounds.shape[0])]
# list of the average parameter updates
sq_para_avg = [0.0 for _ in range(bounds.shape[0])]
# run the gradient descent
for it in range(n_iter):
# calculate gradient
gradient = derivative(solution[0], solution[1])
# update the average of the squared partial derivatives
for i in range(gradient.shape[0]):
# calculate the squared gradient
sg = gradient[i]**2.0
# update the moving average of the squared gradient
sq_grad_avg[i] = (sq_grad_avg[i] * rho) + (sg * (1.0-rho))
# build solution
new_solution = list()
for i in range(solution.shape[0]):
# calculate the step size for this variable
alpha = (ep + sqrt(sq_para_avg[i])) / (ep + sqrt(sq_grad_avg[i]))
# calculate the change
change = alpha * gradient[i]
# update the moving average of squared parameter changes
sq_para_avg[i] = (sq_para_avg[i] * rho) + (change**2.0 * (1.0-rho))
# calculate the new position in this variable
value = solution[i] - change
# store this variable
new_solution.append(value)
# store the new solution
solution = asarray(new_solution)
solutions.append(solution)
# evaluate candidate point
solution_eval = objective(solution[0], solution[1])
# report progress
print('>%d f(%s) = %.5f' % (it, solution, solution_eval))
return solutions
We can then execute the search as before, and this time retrieve the list of solutions instead of the best final solution.
... # seed the pseudo random number generator seed(1) # define range for input bounds = asarray([[-1.0, 1.0], [-1.0, 1.0]]) # define the total iterations n_iter = 120 # rho for adadelta rho = 0.99 # perform the gradient descent search with adadelta solutions = adadelta(objective, derivative, bounds, n_iter, rho)
We can then create a contour plot of the objective function, as before.
... # sample input range uniformly at 0.1 increments xaxis = arange(bounds[0,0], bounds[0,1], 0.1) yaxis = arange(bounds[1,0], bounds[1,1], 0.1) # create a mesh from the axis x, y = meshgrid(xaxis, yaxis) # compute targets results = objective(x, y) # create a filled contour plot with 50 levels and jet color scheme pyplot.contourf(x, y, results, levels=50, cmap='jet')
Finally, we can plot each solution found during the search as a white dot connected by a line.
... # plot the sample as black circles solutions = asarray(solutions) pyplot.plot(solutions[:, 0], solutions[:, 1], '.-', color='w')
Tying this all together, the complete example of performing the Adadelta optimization on the test problem and plotting the results on a contour plot is listed below.
# example of plotting the adadelta search on a contour plot of the test function
from math import sqrt
from numpy import asarray
from numpy import arange
from numpy.random import rand
from numpy.random import seed
from numpy import meshgrid
from matplotlib import pyplot
from mpl_toolkits.mplot3d import Axes3D
# objective function
def objective(x, y):
return x**2.0 + y**2.0
# derivative of objective function
def derivative(x, y):
return asarray([x * 2.0, y * 2.0])
# gradient descent algorithm with adadelta
def adadelta(objective, derivative, bounds, n_iter, rho, ep=1e-3):
# track all solutions
solutions = list()
# generate an initial point
solution = bounds[:, 0] + rand(len(bounds)) * (bounds[:, 1] - bounds[:, 0])
# list of the average square gradients for each variable
sq_grad_avg = [0.0 for _ in range(bounds.shape[0])]
# list of the average parameter updates
sq_para_avg = [0.0 for _ in range(bounds.shape[0])]
# run the gradient descent
for it in range(n_iter):
# calculate gradient
gradient = derivative(solution[0], solution[1])
# update the average of the squared partial derivatives
for i in range(gradient.shape[0]):
# calculate the squared gradient
sg = gradient[i]**2.0
# update the moving average of the squared gradient
sq_grad_avg[i] = (sq_grad_avg[i] * rho) + (sg * (1.0-rho))
# build solution
new_solution = list()
for i in range(solution.shape[0]):
# calculate the step size for this variable
alpha = (ep + sqrt(sq_para_avg[i])) / (ep + sqrt(sq_grad_avg[i]))
# calculate the change
change = alpha * gradient[i]
# update the moving average of squared parameter changes
sq_para_avg[i] = (sq_para_avg[i] * rho) + (change**2.0 * (1.0-rho))
# calculate the new position in this variable
value = solution[i] - change
# store this variable
new_solution.append(value)
# store the new solution
solution = asarray(new_solution)
solutions.append(solution)
# evaluate candidate point
solution_eval = objective(solution[0], solution[1])
# report progress
print('>%d f(%s) = %.5f' % (it, solution, solution_eval))
return solutions
# seed the pseudo random number generator
seed(1)
# define range for input
bounds = asarray([[-1.0, 1.0], [-1.0, 1.0]])
# define the total iterations
n_iter = 120
# rho for adadelta
rho = 0.99
# perform the gradient descent search with adadelta
solutions = adadelta(objective, derivative, bounds, n_iter, rho)
# sample input range uniformly at 0.1 increments
xaxis = arange(bounds[0,0], bounds[0,1], 0.1)
yaxis = arange(bounds[1,0], bounds[1,1], 0.1)
# create a mesh from the axis
x, y = meshgrid(xaxis, yaxis)
# compute targets
results = objective(x, y)
# create a filled contour plot with 50 levels and jet color scheme
pyplot.contourf(x, y, results, levels=50, cmap='jet')
# plot the sample as black circles
solutions = asarray(solutions)
pyplot.plot(solutions[:, 0], solutions[:, 1], '.-', color='w')
# show the plot
pyplot.show()
Running the example performs the search as before, except in this case, the contour plot of the objective function is created.
In this case, we can see that a white dot is shown for each solution found during the search, starting above the optima and progressively getting closer to the optima at the center of the plot.
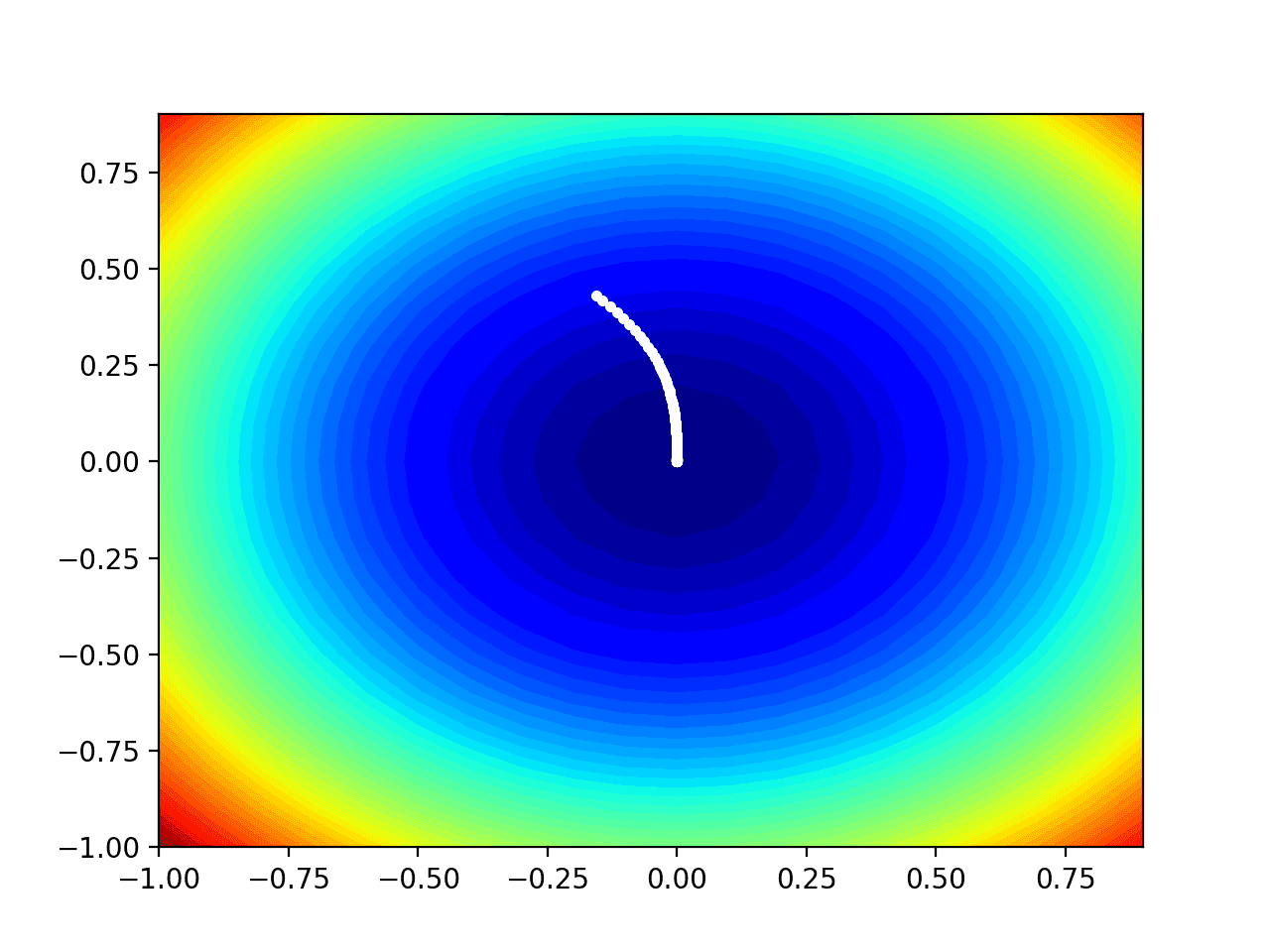
Contour Plot of the Test Objective Function With Adadelta Search Results Shown
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Papers
Books
- Algorithms for Optimization, 2019.
- Deep Learning, 2016.
APIs
Articles
- Gradient descent, Wikipedia.
- Stochastic gradient descent, Wikipedia.
- An overview of gradient descent optimization algorithms, 2016.
Summary
In this tutorial, you discovered how to develop the gradient descent with Adadelta optimization algorithm from scratch.
Specifically, you learned:
- Gradient descent is an optimization algorithm that uses the gradient of the objective function to navigate the search space.
- Gradient descent can be updated to use an automatically adaptive step size for each input variable using a decaying average of partial derivatives, called Adadelta.
- How to implement the Adadelta optimization algorithm from scratch and apply it to an objective function and evaluate the results.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.
The post Gradient Descent With Adadelta from Scratch appeared first on Machine Learning Mastery.
